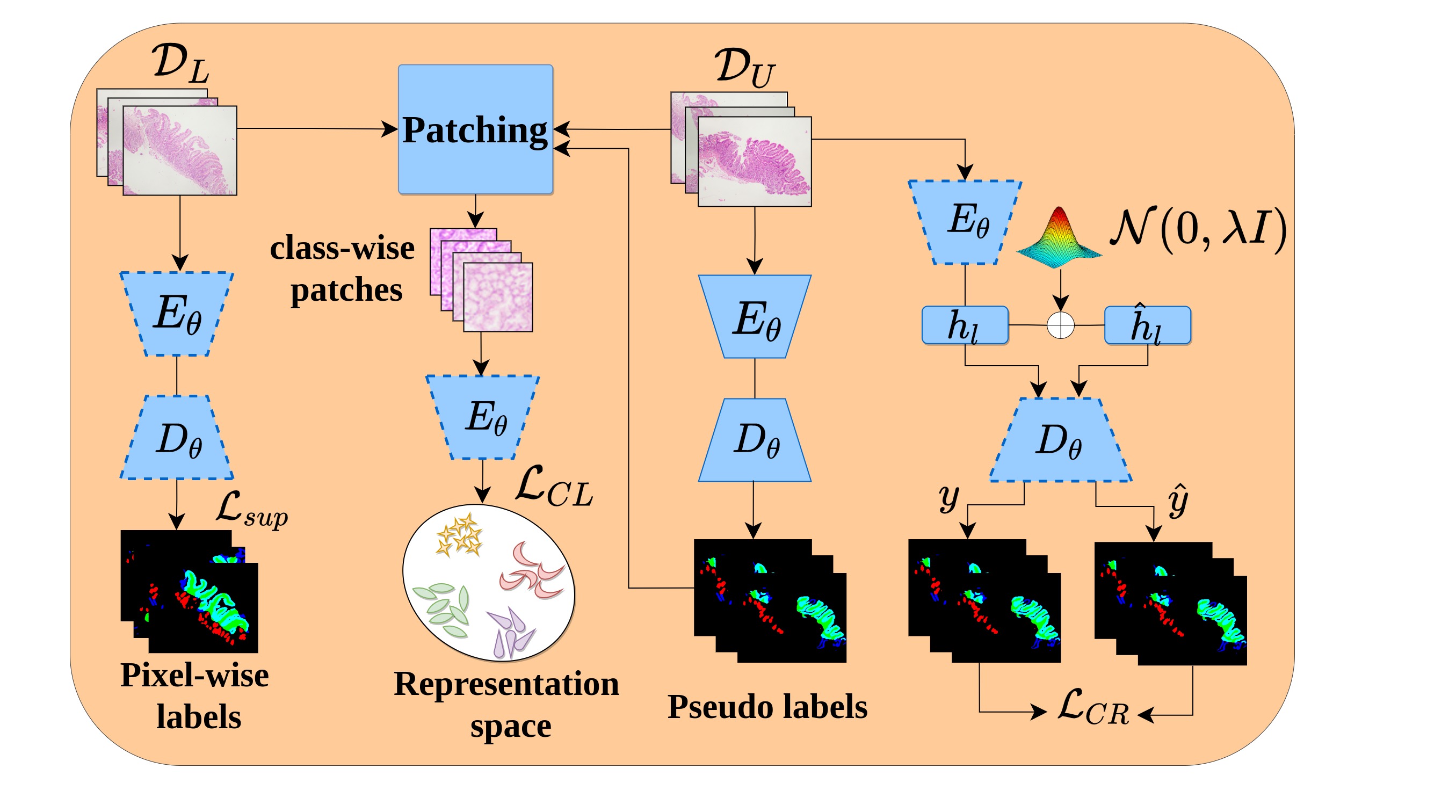
Contrastive Semi-supervised Learning for 2D medical image segmentation.
- attn_unet.py: Code for the backbone/baseline Attention unet
- data_loader.py: Contains data generator for the celiac dataset
- generate_pseudolabels.py: A utility file for generating and saving pseudolabels in case of very slow cpus that cant run on-the-fly
- inference.py: Run inference on a test set of images
- metrics.py: Contains loss functions and metrics to be monitored
- model.py: Defines encoder, decoder, composite model
- main.py: Running semi-supervised CL+CR training and finetuning of the network. Change config variables at the beginning code and run it (python main.py)
- utils.py: Some functional utilities
- dataloaders/data_generator.py: A completely modular implementation of celiac dataset generator. Easily modifiable to other datasets
- FT_generator.py: Contains dataloader for finetuning with Celiac Dataset.
- python 3.6+
- Tensorflow-GPU 2.0+
- Keras (compatible with the TF backend)
Some training images from our duodenal histopathology dataset some pretrained model weights are available here. Each file corresponds to the final weights for the model on that dataset which is used for segmentation. celiac_subset file are sample weights of model trained in supervised manner on a subset (50) of labelled samples only.