Log Neural Controlled Differential Equations (ICML 2024)
[arXiv]
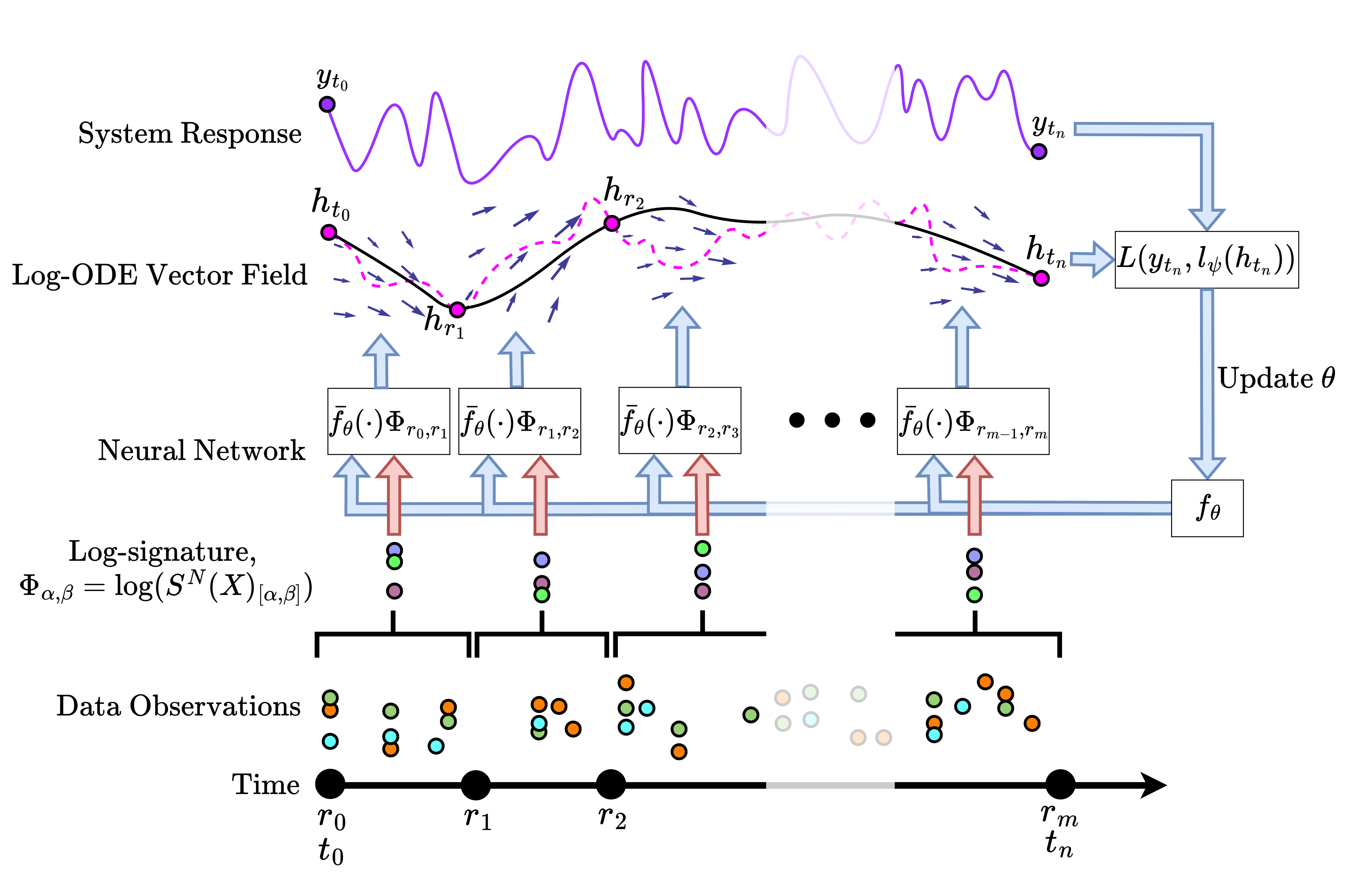
Building on Neural Rough Differential Equations (NRDEs), this repository introduces Log Neural Controlled Differential Equations (Log-NCDEs), a novel, effective, and efficient method for training NCDEs.
Neural controlled differential equations (NCDEs) treat time series data as observations from a control path
Log-NCDEs use the Log-ODE method to approximate the solution path
where
After setting up the JAX environment detailed in the next section, the best place to start is by exploring the simple_example.ipynb notebook.
This Jupyter notebook provides a comprehensive example of training a NCDE and a Log-NCDE on a simple synthetic dataset.
It serves as an example of how to apply the Log-ODE method during NCDE training.
This repository is implemented in python 3.10 and most of the experiments use Jax as their machine learning framework. However, in order to use the efficient Pytorch implementation of the Mamba recurrence, the code for S6 and Mamba is implemented in Pytorch. Although it is possible to install the cuda versions of Jax and Pytorch in the same environment, we recommend using two separate environments. The repository is designed such that the Jax environment is the main environment and the Pytorch environment is only used for the S6 and Mamba experiments.
The code for preprocessing the datasets, training S5, LRU, NCDE, NRDE, and Log-NCDE, and plotting the results uses the following packages:
jaxandjaxlibfor automatic differentiation.equinoxfor constructing neural networks.optaxfor neural network optimisers.diffraxfor differential equation solvers.signaxfor calculating the signature.sktimefor handling time series data in ARFF format.tqdmfor progress bars.matplotlibfor plotting.pre-commitfor code formatting.
conda create -n Log-NCDE python=3.10
conda activate Log-NCDE
conda install pre-commit=3.7.1 sktime=0.30.1 tqdm=4.66.4 matplotlib=3.8.4 -c conda-forge
# Substitue for correct Jax pip install: https://jax.readthedocs.io/en/latest/installation.html
pip install -U "jax[cuda12]" "jaxlib[cuda12]" equinox==0.11.4 optax==0.2.2 diffrax==0.5.1 signax==0.1.1
If running data_dir/process_uea.py throws this error: No module named 'packaging'
Then run: pip install packaging
After installing the requirements, run pre-commit install to install the pre-commit hooks.
The code for training S6 and Mamba uses the following packages:
pytorchfor automatic differentiation.causal-conv1dfor the efficient implementation of a 1D causal convolution.mamba-ssmfor the Mamba layer.einopsfor reshaping tensors.
conda create -n pytorch_mamba python=3.10
conda activate pytorch_mamba
conda install pytorch=2.2.2 pytorch-cuda=12.1 numpy=1.26.4 -c pytorch -c nvidia
conda install packaging=24.1 -c conda-forge
pip install causal-conv1d>=1.2.0 mamba-ssm==1.2.2 einops==0.8.0 jax==0.4.30
The folder data_dir contains the scripts for downloading data, preprocessing the data, and creating dataloaders and
datasets. Raw data should be downloaded into the data_dir/raw folder. Processed data should be saved into the data_dir/processed
folder in the following format:
processed/{collection}/{dataset_name}/data.pkl,
processed/{collection}/{dataset_name}/labels.pkl,
processed/{collection}/{dataset_name}/original_idxs.pkl (if the dataset has original data splits)
where data.pkl and labels.pkl are jnp.arrays with shape (n_samples, n_timesteps, n_features) and (n_samples, n_classes) respectively. If the dataset had original_idxs then those should be saved as a list of jnp.arrays with shape [(n_train,), (n_val,), (n_test,)].
The toy dataset consists of data_dir/toy_dataset.py.
The UEA datasets are a collection of multivariate time series classification benchmarks. They can be downloaded by
running data_dir/download_uea.py and preprocessed by running data_dir/process_uea.py.
The PPG-DaLiA dataset is a multivariate time series regression dataset,
where the aim is to predict a person’s heart rate using data
collected from a wrist-worn device. The dataset can be downloaded from the
UCI Machine Learning Repository. The data should be
unzipped and saved in the data_dir/raw folder in the following format PPG_FieldStudy/S{i}/S{i}.pkl. The data can be
preprocessed by running the process_ppg.py script.
The scripts in the models folder implement a number of deep learning time series models in Jax, including NCDEs,
NRDEs, Log-NCDEs, LRU, and S5. In order to be integrated into the training,
the __call__ function of the model should only take one argument as input. In
order to handle this, the dataloaders return the model's inputs as a list,
which is unpacked within the model __call__.
NCDEs and NRDEs are implemented in models/NeuralCDEs.py. Log-NCDEs are implemented in
models/LogNeuralCDEs.py. The models folder also contains implementations of the following
baseline models:
RNN: A simple recurrent neural network which can use any cell. Currently, the available cells areLinear,GRU,LSTM, andMLP.LRU: A stacked recurrent model with linear recurrent unit layers.S5: A stacked recurrent model with S5 layers.
The torch_experiments folder contains Pytorch implementations of S6 and Mamba. The mamba-ssm package is
used for the mamba recurrence and the S6 recurrence is implemented in torch_experiments/s6_recurrence.py.
The code for training and evaluating the models is contained in train.py for the jax models and
torch_experiments/train.py for the pytorch models. Experiments can be run using the run_experiment.py script. This script requires you to specify the names of the models you want to train,
the names of the datasets you want to train on, and a directory which contains configuration files. By default,
it will run the NCDE, NRDE, Log-NCDE, S5, and LRU experiments. If run with the --pytorch_experiment flag, it will run
the S6 and MAMBA experiments. The configuration files should be organised as config_dir/{model_name}/{dataset_name}.json and contain the
following fields:
seeds: A list of seeds to use for training.data_dir: The directory containing the data.output_parent_dir: The directory to save the output.lr_scheduler: A function which takes the learning rate and returns the new learning rate.num_steps: The number of steps to train for.print_steps: The number of steps between printing the loss.batch_size: The batch size.metric: The metric to use for evaluation.classification: Whether the task is a classification task.lr: The initial learning rate.time: Whether to include time as a channel.- Any specific model parameters.
See experiment_configs/repeats for some examples.
The configuration files for all the experiments with fixed hyperparameters can be found in the experiment_configs folder and
run_experiment.py is currently configured to run the repeat experiments on the UEA datasets for the Jax models.
The results folder contains a zip file of the output files from the UEA, PPG, and toy experiments.
Furthermore, it contains the code for analysing the results and generating the plots in the paper.
When using this code, please cite the following paper:
@inproceddings{Walker2024LogNCDE,
title={Log Neural Controlled Differential Equations: The Lie Brackets Make a Difference},
author={Walker, Benjamin and McLeod, Andrew D. and Qin, Tiexin and Cheng, Yichuan and Li, Haoliang and Lyons, Terry},
journal={International Conference on Machine Learning},
year={2024}
}