This repository contains the suite of tools used in the Hartwig Medical Foundation whole genome,targeted DNA and whole transcriptome analysis pipeline.
Check out what's new.
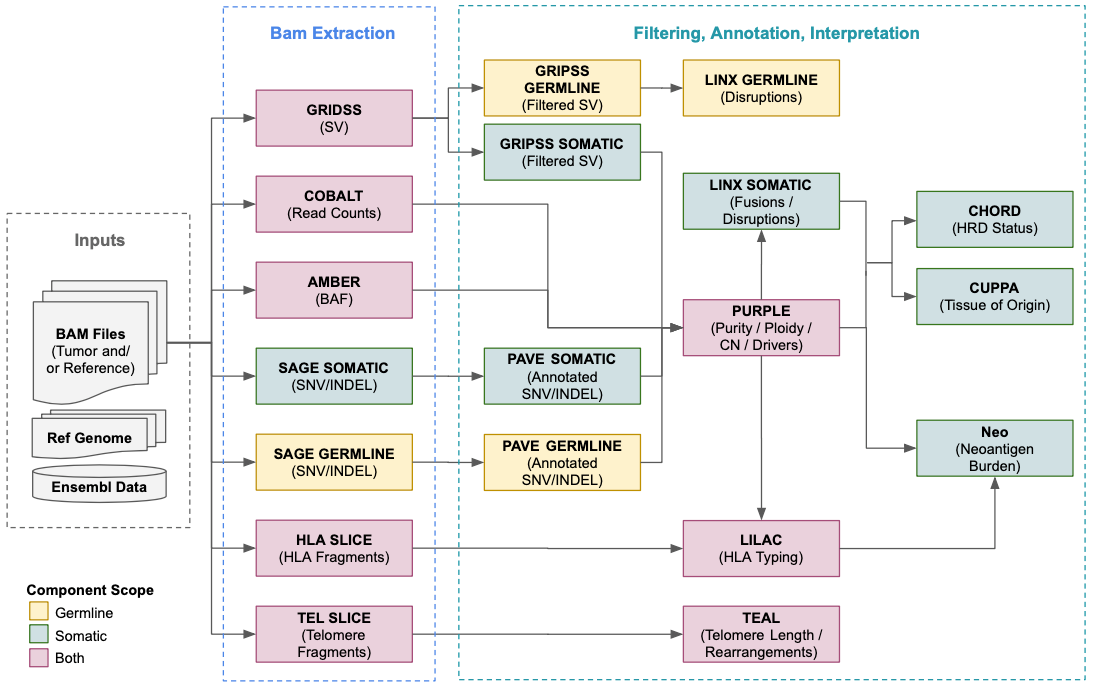
The table below has links for the full functional detail and configuration for each component. The versions match those used in the current HMF GCP pipeline (v5.32) which can be run using Platinum.
| Component | Description | Current Version |
|---|---|---|
| Amber | Generate a tumor BAF file for Purple's copy number fit | 3.9 |
| Cobalt | Determines the read depth ratios for Purple's copy number fit | 1.14 |
| Cuppa | Tissue of origin prediction from WGS/WTS | 1.8 |
| Gripss | SV filtering | 2.3 |
| Lilac | HLA typing | 1.4 |
| Linx | SV annotation, clustering & chaining, fusion and disruption calling | 1.23 |
| Pave | Point mutation annotation and gene impact | 1.4 |
| Purple | Estimates copy number, purity and ploidy, and identifies driver events | 3.8 |
| Sage | Point mutation variant calling and filtering | 3.2 |
| Teal | Measures telomere content and estimates telomeric length | 1.0.1 |
| SvPrep | Pre-GRIDSS BAM filtering | 1.1 |
| Cider | TCR/BCR V(D)J recombination sequence detection | 0.5 |
| BamTools | BAM metrics and slicing | 1.0.1 |
The following external tools are used in the pipeline:
| Component | Description | Current Version |
|---|---|---|
| GRIDSS | Structural variant calling | 2.13.2 |
| Chord | Homologous Recombination Deficiency detection | 2.0 |
An example WGS pipeline which runs each of these components in turn is detailed here. An example targeted-panel pipeline, including support for the HMF and TSO-500 panels, is detailed here.
Resource files for each component (GRCh37 and GRCh38) are available to download from HMFTools-Resources > DNA-Resources.
| Component | Description | Current Version |
|---|---|---|
| Protect | Matching of molecular results to treatments and clinical trials | 2.3 |
| Rose | Actionability of clinically relevant molecular findings | 1.3 |
| Virus Interpreter | Filtering, annotation and interpretation of virus breakend data | 1.2 |
| Orange | PDF summary report and JSON file of all WGS output | 1.10 |
| Patient-reporter | PDF summary report and JSON file of all clinical relevant WGS output | 7.25.1 |
| Component | Description | Current Version |
|---|---|---|
| Isofox | WTS Transcript Abundance, Fusions & Novel Splice Junctions | 1.6 |