DAGMSA: Directed Acyclic Graph-based Multiple Sequence Alignments (MSA) for Protein Multimer Structure Prediction
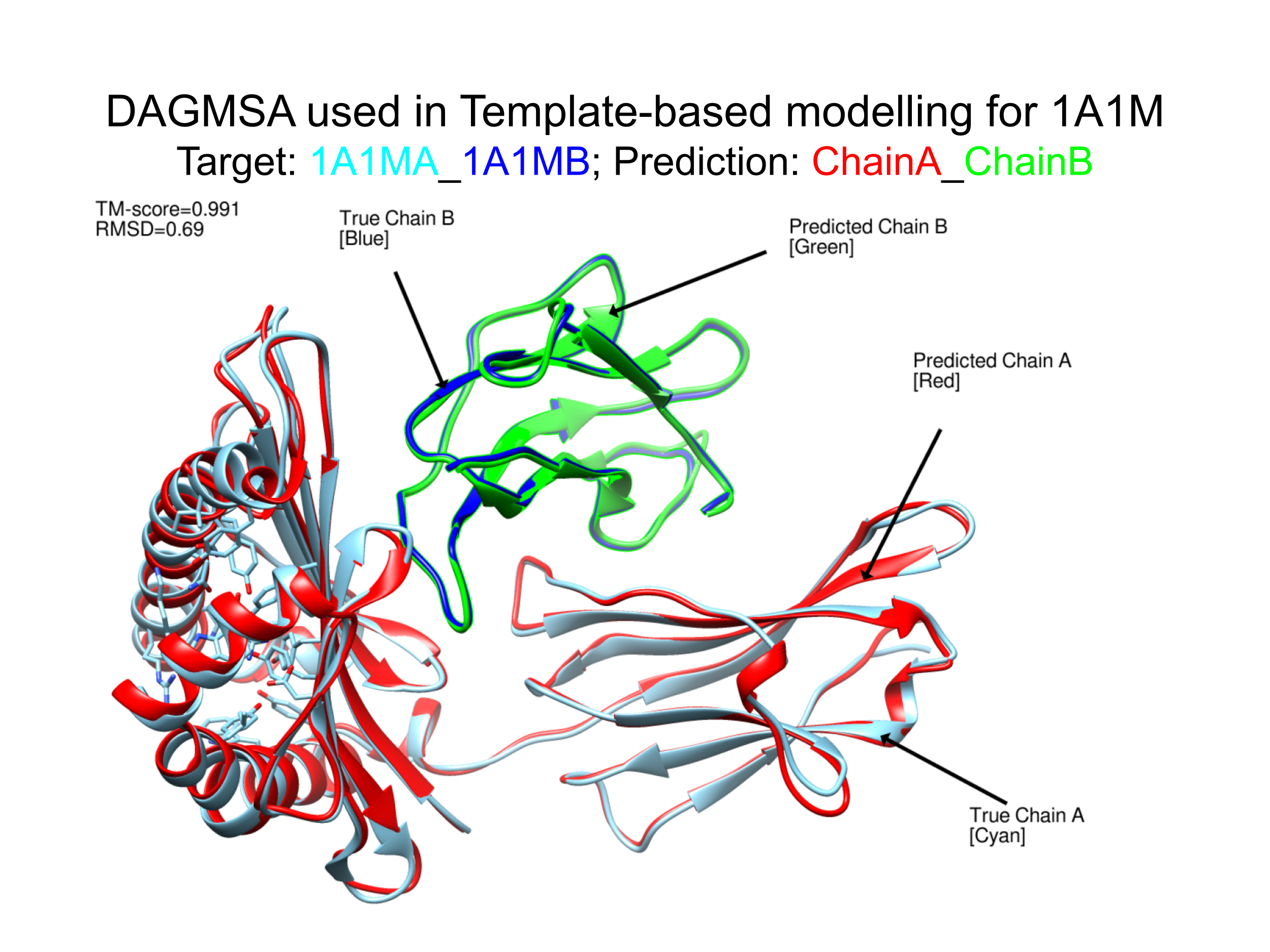
MSA is an important feature of proteins that can illustrate similarities and differences between protein sequences and provide us with co-evolution information which is useful for de novo structure prediction. Although it is relatively easy to obtain MSA of monomeric proteins with many tools available to do so, there are very few tools available for multimeric MSA generation due to the existance of many paralogues and the lack of interacting homologues for higher order multimers. Here we present a new tool called DAGMSA which can be used to generate multiple sequence alignment of higher order oligomers which can complement structure predicting software like Alphafold2-multimer (AF2_M) as well as template-based modelling tools.
Just type the following command to download the software.
git clone https://github.com/BioinfoMachineLearning/dagmsa
You can also use wget to download the software using the following commands:
mkdir dagmsa
cd dagmsa
wget https://github.com/BioinfoMachineLearning/dagmsa
Once the download is complete, follow the steps in the installation section to install and run the software.
If you do not have miniconda already installed then to download and install miniconda just run the following:
sh ./download_install_miniconda.sh
If you already have Miniconda or Anaconda already installed then just run the following to setup the conda environment:
sh ./create_conda_env.sh
The following will install the entire software including all required databases:
sh ./install_dagmsa.sh --db
If you do not wish to download the databases then simply run sh ./install_dagmsa.sh
If you wish to download the databases separately just run sh ./download_db.sh
DAGMSA is now ready to run.
The software takes as input a text file <fasta_list.txt> and an output folder <outdir>.
Please make sure that the <fasta_list.txt> file contains a list of the full paths of the fasta files ordered according to the respective chains. The following is an example:
/path/to/fasta/file/monomer_1.fasta
/path/to/fasta/file/monomer_2.fasta
/path/to/fasta/file/monomer_3.fasta
Run DAGMSA using the following command:
#Usage
sh runDAGMSA.sh <list_file> <output_directory>
#Example
sh runDAGMSA.sh ./example/1A1M_list.txt ./test_1A1M/