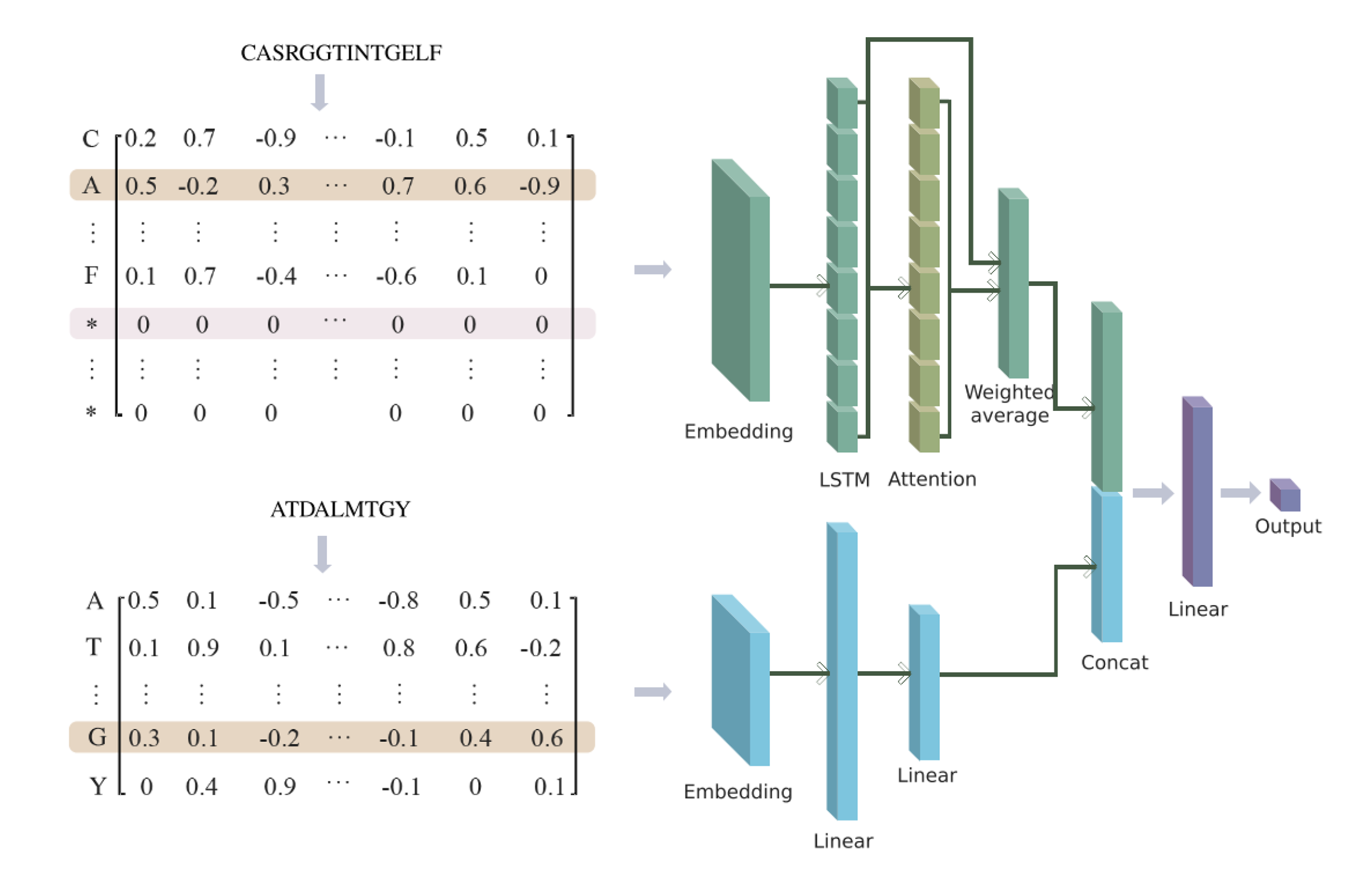
In this study, we developed a dual-input deep learning framework combining bi-directional long short term memory (BiLSTM) and multi-layer perceptron (MLP), named AttnTAP, to extract TCR and peptide features separately and perform TCR-peptide binding prediction.
├── LICENSE <- Non-commercial license.
│
├── README.md <- The README for users using AttnTAP.
│
├── Codes <- Python scripts of AttnTAP. See README for their usages.
│ ├── AttnTAP_test.py <- Making predictions using pre-trained AttnTAP models.
│ ├── AttnTAP_train.py <- Training AttnTAP models.
│ ├── network.py <- The network of AttnTAP.
│ └── AttnTAP_data_clean.py <- Processing raw data files.
│
├── Data <- Data used in AttnTAP.
│ ├── VDJdb
│ │ ├── VDJdb_test.csv
│ │ ├── VDJdb_train.csv
│ │ ├── VDJdb_raw.csv
│ │ └── VDJdb_crossvalid_data <- A set of experimental data for cross-validation.
│ │
│ ├── McPAS
│ │ ├── McPAS_test.csv
│ │ ├── McPAS_train.csv
│ │ ├── McPAS_raw.csv
│ │ └── McPAS_crossvalid_data <- A set of experimental data for cross-validation.
│ │
│ └── example_raw_file.csv <- Example of unprocessed raw data.
│
├── Figure <- Figure used in README.
│ └── AttnTAP.png
│
├── Models <- Pre-trained AttnTAP models for users making predictions directly.
│ ├── cv_model_0_mcpas_0.pth <- The model obtained by taking mcpas_0.csv as the test set and other data as the training set in the cross-validation experiment.
│ └── cv_model_0_vdjdb_0.pth <- The model obtained by taking vdjdb_0.csv as the test set and other data as the training set in the cross-validation experiment.
│
└── Results <- Some results of using AttnTAP.
├── example_data_clean.csv <- The result file after processing `example_raw_file.csv`.
├── cv_0_mcpas_0.csv <- Prediction results on mcpas_0.csv using the corresponding pre-trained model.
└── cv_0_vdjdb_0.csv <- Prediction results on vdjdb_0.csv using the corresponding pre-trained model.
AttnTAP works perfectly in the following versions of the Python packages:
Python 3.9.7
torch 1.11.0+cpu
pandas 1.3.4
numpy 1.20.3
scikit-learn 0.24.2
matplotlib 3.5.1
gensim 4.1.2
Users can use the pre-trained models we provided in ./Models/ to make predictions directly.
First, we need to collect the raw data files, such as ./Data/example_raw_file.csv, set the number of negative samples for each positive sample, such as --neg_samples=1, and use the Python script ./Codes/AttnTAP_data_clean.py to process them by this command:
python ./Codes/AttnTAP_data_clean.py --input_file=./Data/example_raw_file.csv --output_file=./Results/example_data_clean.csv --neg_samples=1
After processing, CDR3 sequences, antigen sequences, and labels, are saved in ./Results/example_data_clean.csv.
tcr antigen label
CASSQEGTGSYEQYF AVFDRKSDAK 1
CASSQEGTGSYEQYF FRDYVDRFYKTLRAEQASQE 0
CATSDRDDSYGYTF AVFDRKSDAK 1
CATSDRDDSYGYTF PKYVKQNTLKLAT 0
CAWSVSGATDEQFF AVFDRKSDAK 1
CAWSVSGATDEQFF RLRAEAQVK 0
Then, we use the Python script ./Codes/AttnTAP_test.py to make predictions and evaluate the performance of AttnTAP on the processed data files in ./Data/McPAS/McPAS_crossvalid_data/0/mcpas_0.csv using the pre-trained model ./Models/cv_model_0_mcpas_0.pth by this command:
python ./Codes/AttnTAP_test.py --input_file=./Data/McPAS/McPAS_crossvalid_data/0/mcpas_0.csv --output_file=./Results/cv_0_mcpas_0.csv --load_model_file=./Models/cv_model_0_mcpas_0.pth
The prediction results, including CDR3 sequences, antigen sequences, and probabilities of binding, are saved in ./Results/cv_0_mcpas_0.csv.
tcr antigen prediction
0 CASSACDCIRKIYGYTF WEDLFCDESLSSPEPPSSSE 0.98302865
1 CASSISSGSYNEQFF VTEHDTLLY 0.5213805
2 CASSSRDIGFYEQYF RAKFKQLL 0.17534448
3 CASSPSGRLNEQYF LPRRSGAAGA 0.9469173
4 CASSPEGTGSYEQYF RPHERNGFTVL 0.4462932
5 CASSPPGIGGYNEQFF RPHERNGFTVL 0.32671145
Accuracy (ACC), area under the receiver operating characteristic curve (AUC), recall (Recall), precision (Precision), F1 score (F1), are calculated and printed as:
-----test results-----
ACC: 0.779
AUC: 0.866
Recall: 0.818
Precision: 0.760
F1: 0.788
The work is done!
Users can use the Python script ./Codes/AttnTAP_train.py to train their own AttnTAP models on their TCR-antigen data samples for a better prediction performance. For example, users can use the processed data file in ./Data/McPAS/McPAS_train.csv to train the model and save the model in ./Models/McPAS_train.pth by this command:
python ./Codes/AttnTAP_train.py --input_file=./Data/McPAS/McPAS_train.csv --save_model_file=./Models/McPAS_train.pth --valid_set=False --epoch=15 --learning_rate=0.005 --dropout_rate=0.1 --embedding_dim=10 --hidden_dim=50 --plot_train_curve=False --plot_roc_curve=False
The training results are calculated and printed as:
Epoch-1 Train loss 0.5603
Epoch-2 Train loss 0.5359
Epoch-3 Train loss 0.5159
Epoch-4 Train loss 0.4941
Epoch-5 Train loss 0.4732
Epoch-6 Train loss 0.452
Epoch-7 Train loss 0.4364
Epoch-8 Train loss 0.4252
Epoch-9 Train loss 0.4035
Epoch-10 Train loss 0.3859
Epoch-11 Train loss 0.3764
Epoch-12 Train loss 0.36
Epoch-13 Train loss 0.3436
Epoch-14 Train loss 0.3255
Epoch-15 Train loss 0.3151
-----train results-----
ACC: 0.875
AUC: 0.951
Recall: 0.885
Precision: 0.867
F1: 0.876
The work is done!
AttnTAP is actively maintained by Xinyang Qian & Fan Li, currently students at Xi'an Jiaotong University in the research group of Prof. Jiayin Wang. If you have any questions, please contact us by e-mail: qianxy@stu.xjtu.edu.cn / lifan0513@stu.xjtu.edu.cn.