Deep MuLti-Instance Learning Improves the Prediction of Cancer-associated T Cell ReceptOrs for Accurate CaNcer Detection
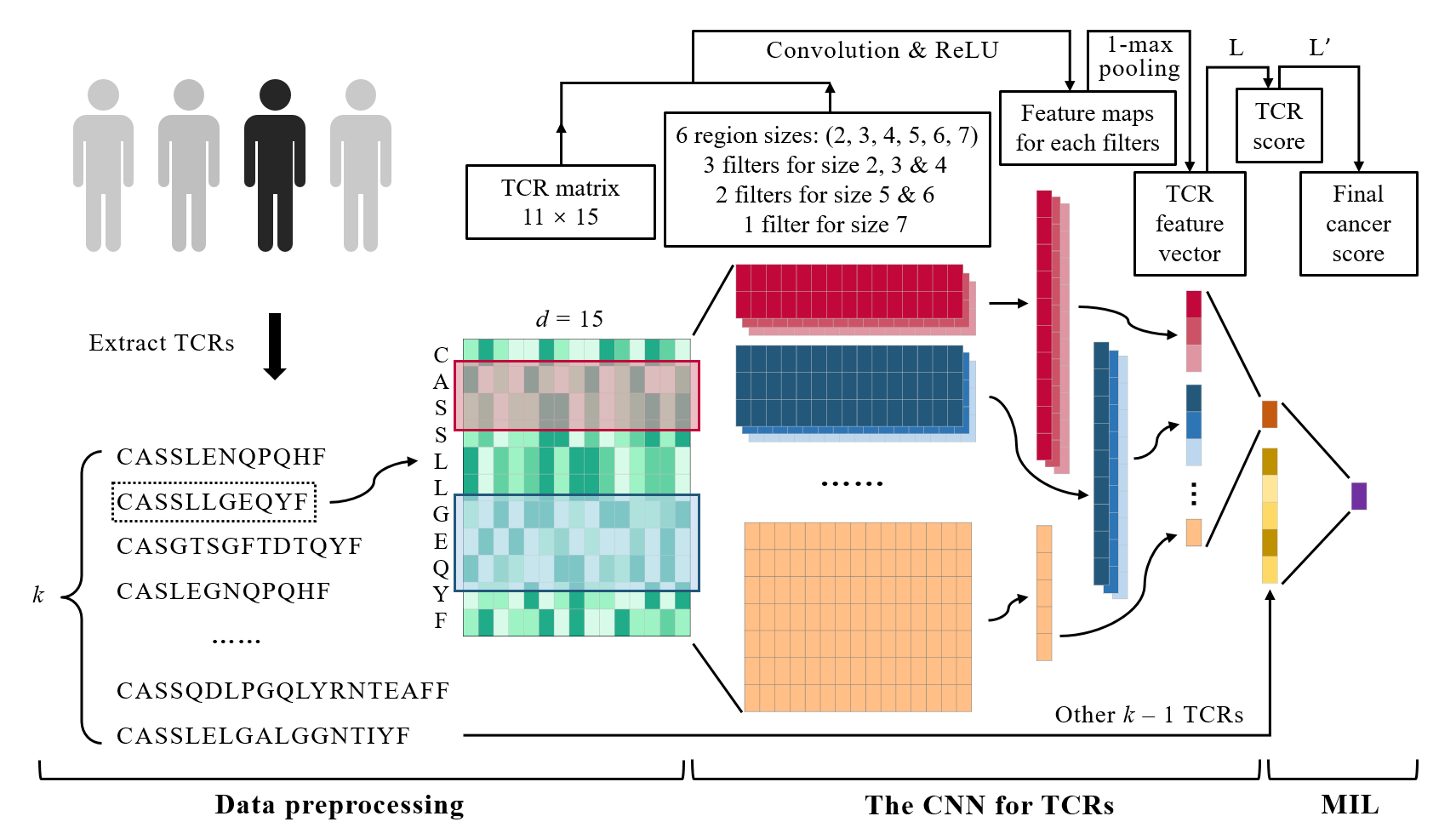
DeepLION is a deep multi-instance learning (MIL) method for identifying cancer-associated T cell receptors (TCRs) and detecting cancer samples using TCR-sequencing data. Its workflow is divided into three parts: data preprocessing, the convolutional neural network (CNN) for TCRs, and MIL. For more details, please read our paper DeepLION: Deep Multi-Instance Learning Improves the Prediction of Cancer-Associated T Cell Receptors for Accurate Cancer Detection.
├── LICENSE <- Non-commercial license.
│
├── README.md <- The top-level README for users using DeepLION.
│
├── Codes <- Python scripts of DeepLION. See README for their usages.
│ ├── DeepLION_prediction.py <- Making predictions using pre-trained DeepLION models.
│ ├── DeepLION_training.py <- Training DeepLION models.
│ ├── Evaluation.py <- Evaluating the performances of DeepLION.
│ └── ProcessRawFiles.py <- Processing raw TCR-sequencing data files.
│
├── Data <- Data used in DeepLION. See README in this folder for more details.
│ ├── Lung
│ │ ├── TestData
│ │ ├── TrainingData
│ │ └── README.md
│ │
│ ├── THCA
│ │ ├── TestData
│ │ ├── TrainingData
│ │ └── README.md
│ │
│ ├── TrainingSequences
│ │ ├── NormalCDR3.txt
│ │ ├── NormalCDR3_test.txt
│ │ ├── README.md
│ │ ├── TumorCDR3.txt
│ │ └── TumorCDR3_test.txt
│ │
│ ├── AAidx_PCA.txt
│ ├── Example_raw_file.tsv
│ ├── README.md
│ └── Reference_dataset.tsv
│
├── Figures <- Figures used in README.
│ ├── DeepLION_workflow.png
│ └── Lion.png
│
├── Models <- Pre-trained DeepLION models for users making predictions directly.
│ ├── Pretrained_Lung.pth
│ └── Pretrained_THCA.pth
│
└── Results <- Some results of using DeepLION.
├── Example.tsv <- The result file after processing `Example_raw_file.tsv`.
├── Lung_prediction.tsv <- Prediction results on lung cancer test data using the corresponding pre-trained model.
└── THCA_prediction.tsv <- Prediction results on thyroid cancer test data using the corresponding pre-trained model.
DeepLION works perfectly in the following versions of the Python packages:
Python 3.7.2
numpy 1.21.2
torch 1.6.0+cpu
scikit-learn 0.23.2
Users can use the pre-trained models we provided in ./Models/Pretrained/ to make predictions directly.
First, we need to collect the raw TCR-sequencing data files, such as ./Data/Example_raw_file.tsv, and use the Python script ./Codes/ProcessRawFiles.py to process them by this command:
python ./Codes/ProcessRawFiles.py --input ./Data/Example_raw_file.tsv --reference ./Data/Reference_dataset.tsv --output ./Results/Example.tsv
After processing, the low-quality TCR beta chain CDR3 sequences and the sequences appearing in the reference dataset are removed. The top k (default: 100) TCR sequences and their abundances are saved in ./Results/Example.tsv:
TCR Abundance
CASSLTRLGVYGYTF 0.06351
CASSKREIHPTQYF 0.043778
CASSLEGGAAMGEKLFF 0.039882
CASSPPDRGAFF 0.034422
CASSTGTAQYF 0.028211
CASSEALQNYGYTF 0.027918
CSARADRGQGYEQYF 0.027427
CASSPWAATNEKLFF 0.023224
CAWGWTGGTYEQYF 0.019363
······
If users get raw files in different format, they can also apply this script by setting the argument --info_index (default: [-3. 2]) to the indexes of CDR3 sequences and their clone fractions in their files.
Then, we use the Python script ./Codes/DeepLION_prediction.py to make predictions on processed data files in ./Data/THCA/TestData/ using the pre-trained model ./Models/Pretrained_THCA.pth by this command:
python ./Codes/DeepLION_prediction.py --sample_dir ./Data/THCA/TestData/ --model_file ./Models/Pretrained_THCA.pth --aa_file ./Data/AAidx_PCA.txt --output ./Results/THCA_prediction.tsv
The prediction results, including sample filenames, probabilities of being cancer-associated, and cancer predictions, are saved in ./Results/THCA_prediction.tsv:
Sample Probability Prediction
Health_001.tsv 0.2733137767311635 False
Health_002.tsv 0.11589630679459391 False
Health_003.tsv 0.0023036408351775795 False
Health_004.tsv 0.04514491246460731 False
Health_005.tsv 0.03503014357993675 False
Health_006.tsv 0.0008458254917743452 False
Health_007.tsv 0.26301584490197166 False
Health_008.tsv 0.04840260793287661 False
Health_009.tsv 0.00024520156538897236 False
······
Finally, we can use the Python script ./Codes/Evaluation.py to evaluate the performance of DeepLION on these test data by this command:
python ./Codes/Evaluation.py --input ./Results/THCA_prediction.tsv
The metrics, accuracy, sensitivity, specificity, and area under the receiver operating characteristic (ROC) curve (AUC), are calculated and printed as:
----- [./Results/THCA_prediction.tsv] -----
Accuracy: 0.884
Sensitivity: 0.8
Specificity: 0.957
AUC: 0.956
Users can use the Python script ./Codes/DeepLION_training.py to train their own DeepLION models on their TCR-sequencing data samples for a better prediction performance by this command:
python ./Codes/DeepLION_training.py --sample_dir ./Data/THCA/TrainingData/ --aa_file ./Data/AAidx_PCA.txt --dropout 0.4 --epoch 1000 --learning_rate 0.001 --output ./Models/Pretrained_THCA.pth
When using our results or modelling approach in a publication, please cite our paper (https://doi.org/10.3389/fgene.2022.860510):
Xu Y, Qian X, Zhang X, Lai X, Liu Y and Wang J (2022) DeepLION: Deep Multi-Instance Learning Improves the Prediction of Cancer-Associated T Cell Receptors for Accurate Cancer Detection. Front. Genet. 13:860510. doi: 10.3389/fgene.2022.860510
DeepLION is actively maintained by Xinyang Qian, currently a Ph.D student at Xi'an Jiaotong University in the research group of Prof. Jiayin Wang.
If you have any questions, please contact us by e-mail: qianxy@stu.xjtu.edu.cn.