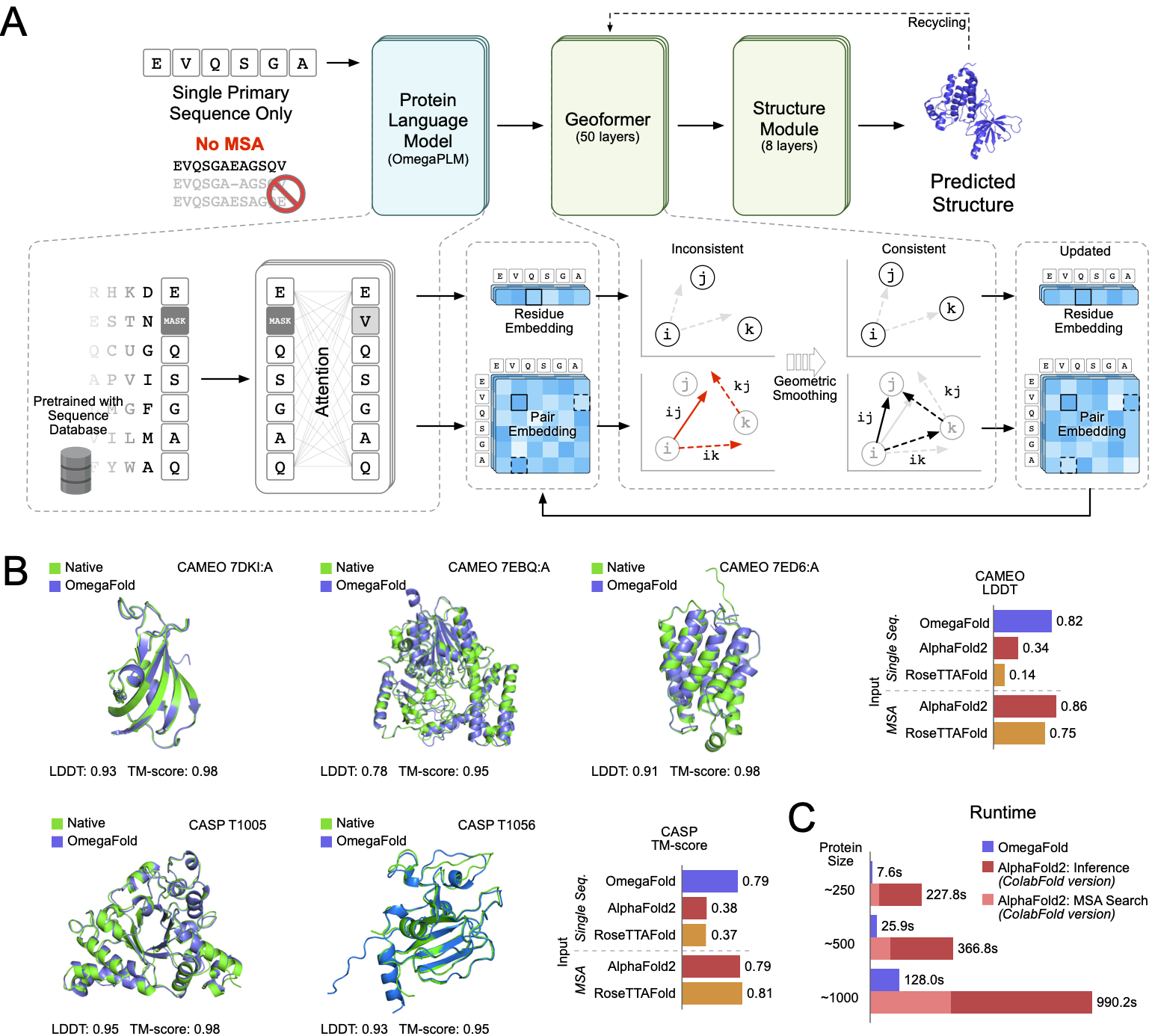
This is the release code for paper High-resolution de novo structure prediction from primary sequence.
We will continue to optimize this repository for more ease of use, for instance, reducing the GRAM required to inference long proteins and releasing possibly stronger models.
Now you can use model 2 by setting --model 2 in the command line!
We have optimized (to some extent) the GRAM usage of OmegaFold model in our
latest release. Now the model can inference protein sequence as long as
4096 on NVIDIA A100 Graphics card with 80 GB of memory with
--subbatch_size set to 448 without hitting full memory.
This version's model is more sensitive to --subbatch_size.
Subbatch makes a trade-off between time and space.
One can greatly reduce the space requirements by setting --subbatch_size
very low.
The default is the number of residues in the sequence and the lowest
possible number is 1.
For now we do not have a rule of thumb for setting the --subbatch_size,
but we suggest half the value if you run into GPU memory limitations.
For macOS users, we support MPS (Apple Silicon) acceleration if the user
installs the latest nightly version of PyTorch.
Also, current code also requires macOS users need to git clone the
repository and use python main. py (see below) to run the model.
To prepare the environment to run OmegaFold,
- from source
pip install git+https://github.com/Bo-Ni/OmegaFold_0.git
- clone the repository
git clone https://github.com/HeliXonProtein/OmegaFold
cd OmegaFold
python setup.py install
should get you where you want.
The INPUT_FILE.fasta should be a normal fasta file with possibly many
sequences with a comment line starting with > or : above the amino
acid sequence itself.
This command will download the weight
from https://helixon.s3.amazonaws.com/release1.pt
to ~/.cache/omegafold_ckpt/model.pt
and load the model
You could simply
omegafold INPUT_FILE.fasta OUTPUT_DIRECTORY
And voila!
Even if this failed, since we use minimal 3rd party libraries, you can always just install the latest PyTorch and biopython (and that's it!) yourself. For mps accelerator, macOS users may need to install the lastest nightly version of PyTorch. In this case, you could run
python main.py INPUT_FILE.fasta OUTPUT_DIRECTORY
However, since we have implemented sharded execution, it is possible to
- trade computation time for GRAM: by changing
--subbatch_size. The smaller this value is, the longer the execution can take, and the less memory is required, or, - trade computation time for average prediction quality, by changing
--num_cycle
For more information, run
omegafold --help
where we provide several options for both speed and weights utilities.
We produce one pdb for each of the sequences in INPUT_FILE.fasta saved in
the OUTPUT_DIRECTORY. We also put our confidence value the place of
b_factors in pdb files.
If this is helpful to you, please consider citing the paper with
@article{OmegaFold,
author = {Wu, Ruidong and Ding, Fan and Wang, Rui and Shen, Rui and Zhang, Xiwen and Luo, Shitong and Su, Chenpeng and Wu, Zuofan and Xie, Qi and Berger, Bonnie and Ma, Jianzhu and Peng, Jian},
title = {High-resolution de novo structure prediction from primary sequence},
elocation-id = {2022.07.21.500999},
year = {2022},
doi = {10.1101/2022.07.21.500999},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2022/07/22/2022.07.21.500999},
eprint = {https://www.biorxiv.org/content/early/2022/07/22/2022.07.21.500999.full.pdf},
journal = {bioRxiv}
}
Also some of the comments might be out-of-date as of now, and will be updated very soon