Tame your model output with INTerval-bAsed Marginal Effects.
Install the development version from GitHub (using devtools):
devtools::install_github("bodoburger/intame")library(intame)
library(nnet)
set.seed(4219)
n = 200
x = runif(n, min = 0, max = 1)
x1 = runif(n, min=0, max=1) + .5*x
x2 = runif(n, min=0, max=1) + .5*x
y2 = function(x) -4 * cos(4*pi*x) * x + 4
y = 4*x1 + y2(x2) + rnorm(n, 0, .5)
df = data.frame(y, x1, x2)
nnet.mod = nnet(y ~ ., df, size = 20, linout = TRUE,
decay = 0.0001, maxit = 1000, trace = FALSE)
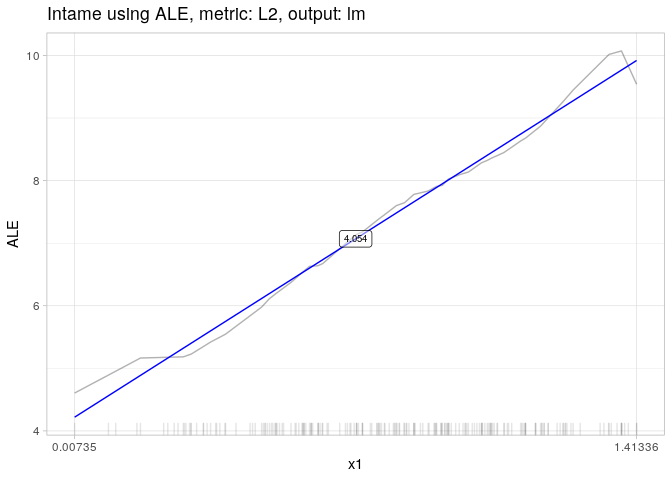
AME.x1 = intame(nnet.mod, df, "x1")
AME.x1## [0.007, 1.413)
## 4.054118
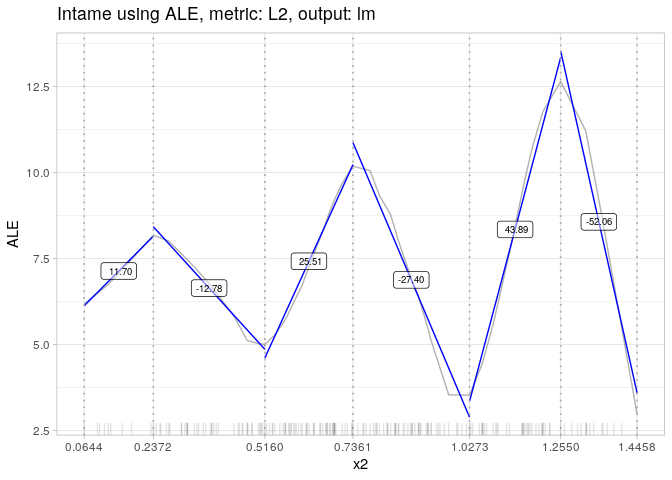
plot(AME.x1)AME.x2 = intame(nnet.mod, df, "x2")
AME.x2## [0.064, 0.237) [0.237, 0.516) [0.516, 0.736) [0.736, 1.027) [1.027, 1.255)
## 11.69899 -12.78246 25.51410 -27.39529 43.89440
## [1.255, 1.446]
## -52.06151
plot(AME.x2)See Rmarkdown files.