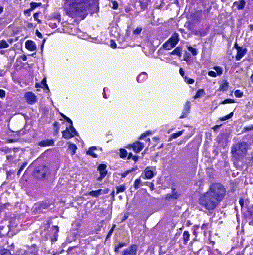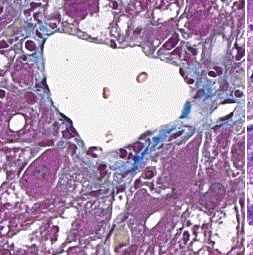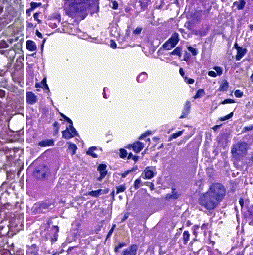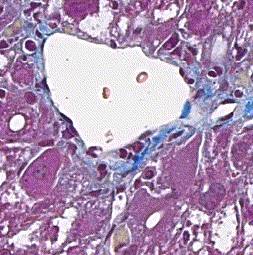
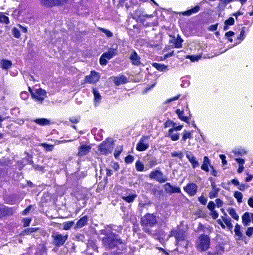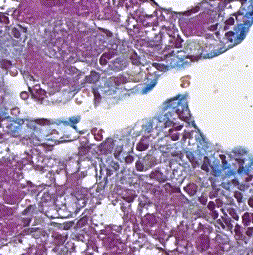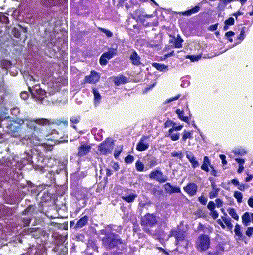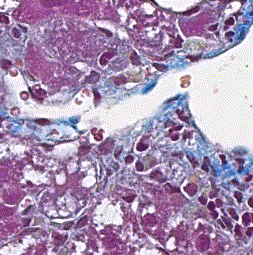
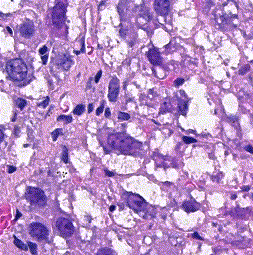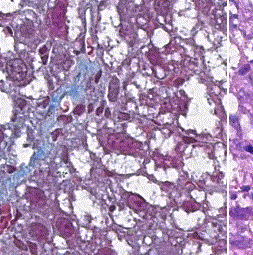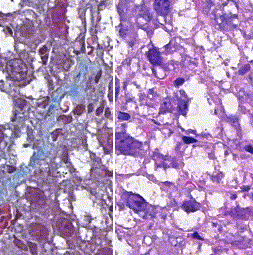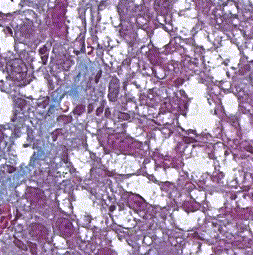
This repository accompanies Zingman et al. A comparative evaluation of image-to-image translation methods for stain transfer in histopathology (MIDL, 2023). The paper analyses strengths and weaknesses of image-to-image translation methods for stain transfer in histopathology, thereby allowing a rational choice of the most suitable approach.
├── README.md <- The top-level README for developers using this project.
│
├── config.yaml <- Default yaml configuration file for inference experiments.
│
├── data <- Directory for input data.
│
├── models <- Trained and serialized models.
│
├── results <- Generated images and metrics are saved here.
│
├── requirements.txt <- The requirements file for reproducing the analysis environment, e.g. generated with `pip freeze > requirements.txt`
│
├── src <- Source code for use in this project.
│ │
│ ├── data_processing <- Data manipulation functionalities.
│ │
│ ├── modelling <- Model arquitectures and definitions.
│ │
│ ├── tests <- Tests directory.
│ │
│ └── utils <- Utilities used by other scripts.
│
├── main.py <- Main execution file, used for generating fake images and for calculating metrics.
│
├── metric_calculation.py <- Calculates FID, WD, and SSIM metrics for generated images
│
└── visualizer.py <- Tool for visualizing generated fake images and inspecting metrics.
- Linux or macOS
- Python 3.7 +
- CPU or NVIDIA GPU + CUDA CuDNN
Create a working environment, with e.g. conda and activate it. It is recommended not to use virtualenv as there are problems with the spams library.
Clone this repository: git clone https://github.com/Boehringer-Ingelheim/stain-transfer
Install spams: conda install -c conda-forge python-spams.
Install the required modules pip install -r requirements.txt.
Download trained models and test or validation histopathological dataset from https://osf.io/byf27/.
Trained models can be saved in models/ folder, H&E stained samples and Masson's trichrome stained samples
from the dataset can be saved in data/he/ and data/masson/ folders.
- Run
python main.py --conf config_example_he2mt.yaml, or use a different yaml configuration file avaialble in the root folder of the project. This will generate artificially created samples (images with Masson's Trichrome artificially stained tissue from images of H&E stained tissue). The detailed description of the fields in the configuration file is available inconfig.yaml - Run
python metric_calculation.py --real_source data/he/ --real_target data/masson/ --fakes results/masson_fake/. The given paths correspond to the paths inconfig_example_he2mt.yaml. This will generate excel file with computed FID, WD, and SSIM metrics that evaluate the quality of the created images of artificially stained tissue.
All parameters for generating fakes are inside the generate key in the yaml.
You will find a models key here which is for specifying what models to use,
and their associated weights. If weights are not provided, then default
weights for each model will be loaded, based on the a2b key which specifies
the direction of domain translation. The default weights are located in the
models folder and the name of each file is model_direction.pth where model
is the name of the model in lowercase and direction is either he2mt or _
mt2he_. For example, each of the following configuration would be using the same
model and weights file:
generate:
models:
names: [ cyclegan ]
weights: [ models/cyclegan_he2mt.pth ]
a2b: true |
generate:
models:
names: [ cyclegan ]
a2b: true |
generate:
models:
names: [ cyclegan ]
weights: [ retrained.pth ]
a2b: trueYou can generate fakes for a given data_path using multiple models at once:
generate:
models:
names: [ cyclegan, cut, munit, macenko, pix2pix, vahadane ]
a2b: true
data_path: data/processed/HE/Available models are:
- colorstat
- cut
- cyclegan
- drit
- macenko
- munit
- pix2pix
- staingan
- stainnet
- unit
- utom
- vahadane
For munit, drit, colorstat, macenko and vahadane there are different inference configurations available.
For munit and drit (the higher the number the higher the precedence):
- The default mode is to generate a random style/attribute tensor (munit/drit) and use it when translating between domains.
- Instead of a random style/attribute tensor (munit/drit), you can provide a
specific precomputed one by setting the path to such tensor in
the
target_tensorkey. This precomputed tensor can be for example, the average style/attribute of all images in the target domain. - You can also compute style/attribute tensors (munit/drit) on the fly for a
given set of images by providing the path to the images in
target_path. The number of target images used for each translation will be the same asbatch_size.
For colorstat, macenko and vahadane (the higher the number the higher the precedence):
- The default mode is to use default "weights". These "weights" are actually
tensors representing averages of all images in our training set for each
domain. These averages are:
- means and standard deviations for colorstat
- stain matrix and 99th percentile of concentration matrix for macenko
- stain matrix and 99th percentile of concentration matrix for vahadane
- By providing the path to a specific precomputed tensor in
the
target_tensorkey. - By computing average means and standard deviations or stain matrices and
corresponding 99th percentiles of concentration matrices on the
fly for a give set of images by providing the path to the images
in
target_pathand the number of images to be considered in each translation intarget_samples.
You can use inference modes 1. and 3. (default mode and computing tensors on the fly) with multiple of these models, and other models, from a single configuration:
generate:
models:
names: [ cyclegan, cut, munit, macenko, pix2pix, vahadane ]
a2b: true
data_path: data/processed/HE/
target_path: data/processed/masson_trichrome
target_samples: 2If you are using inference mode 2. (precomputed tensors) for any of these models, then you can't generate fakes at once from one single configuration file that includes any two of these models. The configuration on left is wrong, since this target tensor is a style tensor for munit, and would fail for macenko and vahadane. The one on the right is valid.
generate:
models:
names: [ cut, munit, macenko, pix2pix, vahadane ]
a2b: true
data_path: data/processed/HE/
target_tensor: precomputed_munit_style_tensor.pth |
generate:
models:
names: [ cut, munit, pix2pix ]
a2b: true
data_path: data/processed/HE/
target_tensor: precomputed_munit_style_tensor.pth |
There is also the option to precompute average tensors for all images in a given path. To do so, specify one of the following models in the configuration yaml:
- colorstataveragetensor: for the given
data_pathcomputes the average mean and std of images. - macenkoaveragetensor: for the given
data_pathcomputes the average stain matrix and 99th percentile of the concentration matrix of images using macenko method. - vahadaneaveragetensor: same as macenkoaveragetensor but with vahadane method.
- munitaveragetensor: for the given
data_pathcomputes the average style of images. The default munit weights will be used ifweightsis not specified in the configuration. When using this modela2bkey should be specified to know ifdata_pathcontains images from domain A or B. - dritaveragetensor: same as munitaveragetensor, only that instead of styles, attributes are computed.
Computed average tensors will be saved in results/av_tensors folder if no
results_path is provided in the configuration. Each model will have its own
sub-folder with computed tensors. The computed tensors can then be set
as target_tensor when generating fakes.
Use metric_calculation.py to compute the metrics, e.g.:
python metric_calculation.py --real_source data/he/ --real_target data/masson/ --fakes results/masson_fake/ --device 0
Provide the following required arguments:
- real_source: path to real source images (for SSIM)
- real_target: path to real target images (for FID, WD)
- fakes: path to folder containing folders with fake images
A csv with SSIM, FID and WD will be generated.
Performance of different Image-to-Image translation methods on validation dataset (please, see the details in A comparative evaluation of image-to-image translation methods for stain transfer in histopathology).
| Model | FID |
WD |
SSIM |
|---|---|---|---|
| CycleGAN | 16.33 | 1.46 | 0.951 |
| CUT | 17.10 | 1.60 | 0.914 |
| MUNIT | 19.20 | 1.61 | 0.871 |
| StainGAN | 19.59 | 3.27 | 0.952 |
| UNIT | 20.23 | 2.54 | 0.940 |
| UTOM | 20.64 | 2.32 | 0.952 |
| DRIT | 22.83 | 2.06 | 0.915 |
| Pix2Pix | 48.47 | 8.42 | 0.998 |
| StainNet | 50.49 | 11.41 | 0.972 |
| ColorStat | 62.13 | 9.60 | 0.974 |
| Macenko | 70.39 | 12.90 | 0.926 |
| Vahadane | 76.55 | 15.14 | 0.911 |
@inproceedings{zingman2024comparative,
title={A comparative evaluation of image-to-image translation methods for stain transfer in histopathology},
author={Zingman, Igor and Frayle, Sergio and Tankoyeu, Ivan and Sukhanov, Sergey and Heinemann, Fabian},
booktitle={Medical Imaging with Deep Learning},
pages={1509--1525},
year={2024},
organization={PMLR}
}