The GA4GH Variation Representation Specification and accompanying specification aim to provide a comprehensive coverage for all types of biological sequence variation.
Specific goals for the project
- Develop common language- and protocol-neutral information models and nomenclature for biological sequence variation.
- From the information models, develop data schemas. The current schema is defined in JSON Schema, but other formats are expected.
- Provide algorithmic guidance and conventions to minimize representational ambiguity.
- Define a globally unique computed identifier for all variation types.
- Develop a reference implementation.
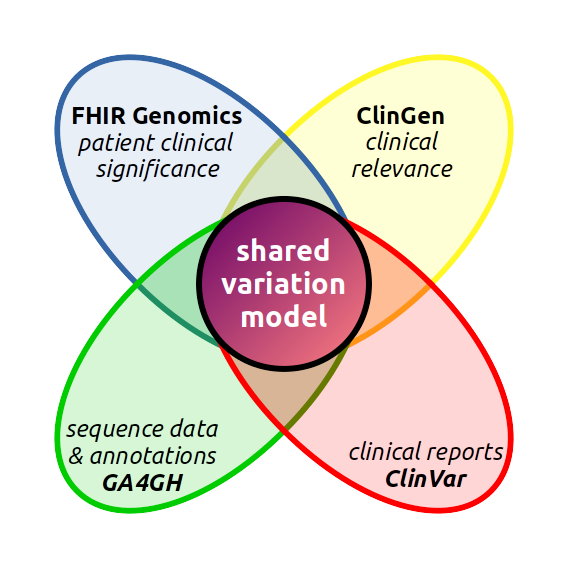
The VR model is the product of the GA4GH Variation Representation group.
The genesis of the GA4GH VR project was the Variation Modelling Collaboration (VMC), which was formed as an independent collaboration of members of ClinGen, ClinVar, FHIR, and GA4GH. The VMC specification received many comments and encouragements, but ultimately was difficult to use. In 2017, the VMC group was absorbed into the GA4GH and underwent a major membership change. In 2019, the effort is undergoing a major overhaul to adapt ideas from VMC to a new and more usable model.
- Gil Alterovitz, Harvard Medical School/Boston Children’s Hospital, FHIR Genomics
- Larry Babb, Sunquest, ClinGen
- Karen Eilbeck, University of Utah
- Bob Freimuth, Mayo Clinic, ClinGen, HL7/FHIR
- Reece Hart, Invitae, GA4GH, chair
- Sarah Hunt, Ensembl
- David Kreda, Harvard Medical School, FHIR Genomics
- Jennifer Lee, NCBI, ClinVar
- Peter Robinson, Jackson Laboratory, HPO
- Shawn Rynearson, University of Utah, UCGD
VMC received significant support from the Broad Institute, GA4GH, and Invitae. Current (2019) developments in the model are supported by GA4GH and GA4GH driver projects.
- VMC Data Model and Specifications (DRAFT) This document is stale. It will be overhauled to match the current schema.
- Google Drive folder
- vmc-discuss mailing list