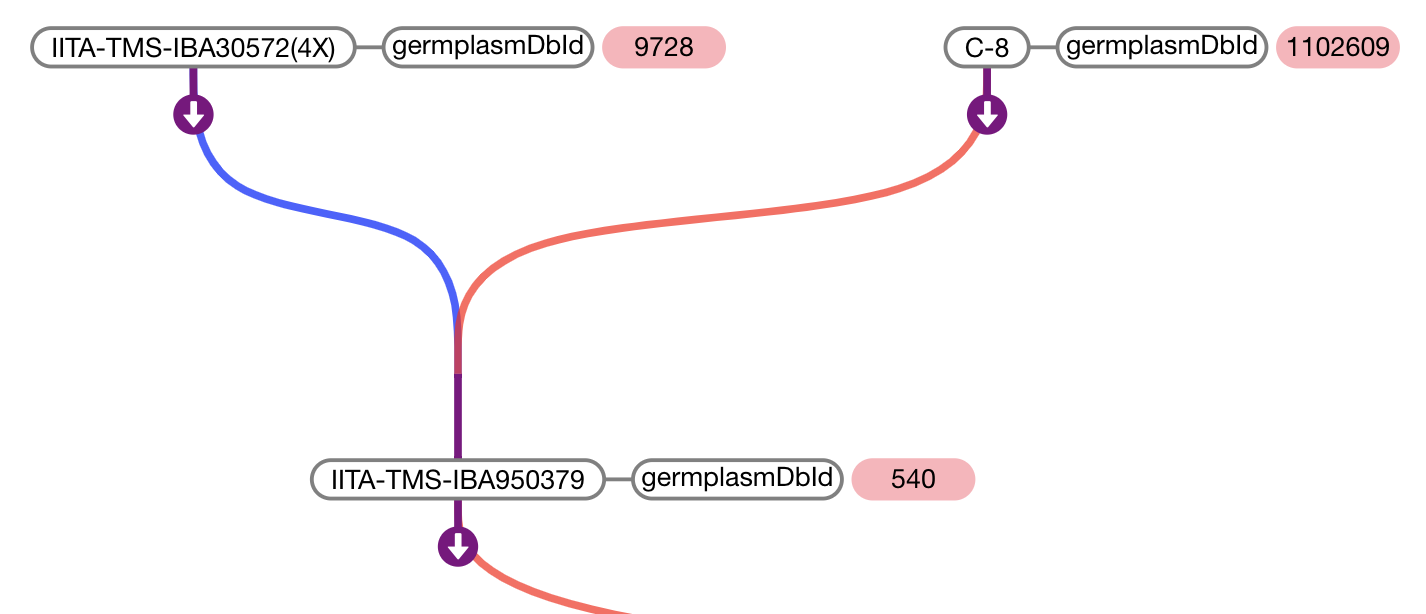
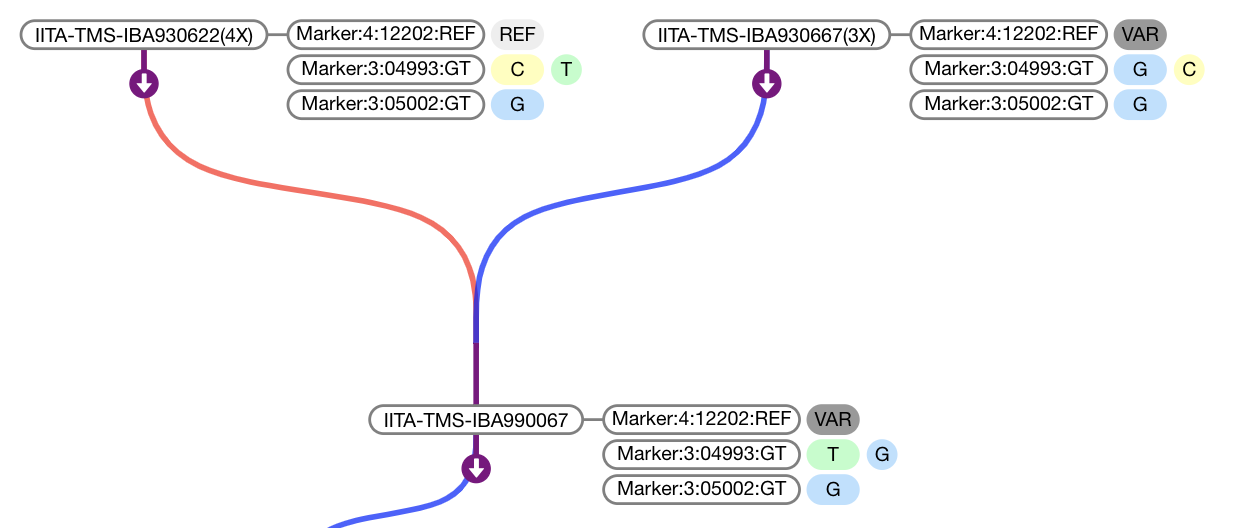
BrAPI Pedigree Viewer 

- Try it out here
- Include-ready version can be found under releases.
- Also available via npm:
npm install @solgenomics/brapi-pedigree-viewer
The pedigree viewer exposes a minimal API for initializing and populating the tree. It relies on BrAPI to fetch the pedigree data it uses.
Use the following snippets (in order) to fully set up a tree.
- Create a new PedigreeTree object (create one for each pedigree tree viewer)
var pdg = PedigreeViewer( //REQUIRED Server: BrAPI.js handle for target endpoint BrAPI("https://brapi.myserver.org/brapi/v1"), //OPTIONAL a function which returns a link to a germplasm information page, returning null will create a node without a link. function(germplasmDbId){ return "https://brapi.myserver.org/germ/"+germplasmDbId+".html"; }, //OPTIONAL additional configuration options { credentials: 'same-site', //see https://developer.mozilla.org/en-US/docs/Web/API/Request/credentials for options urlTarget: '_blank', //see https://developer.mozilla.org/en-US/docs/Web/HTML/Element/a#attr-target for options nodeNameFn: function(d) { // allows for customization of the name displayed on a pedigree node return d.value.name; }, textSize: "14", textFont: "sans-serif", arrowRight: function() { //resolve the right arrow display return ""; }, arrowUp: function() { //resolve the up arrow display return ""; }, arrowDown: function() { //resolve the down arrow display return ""; }, } );
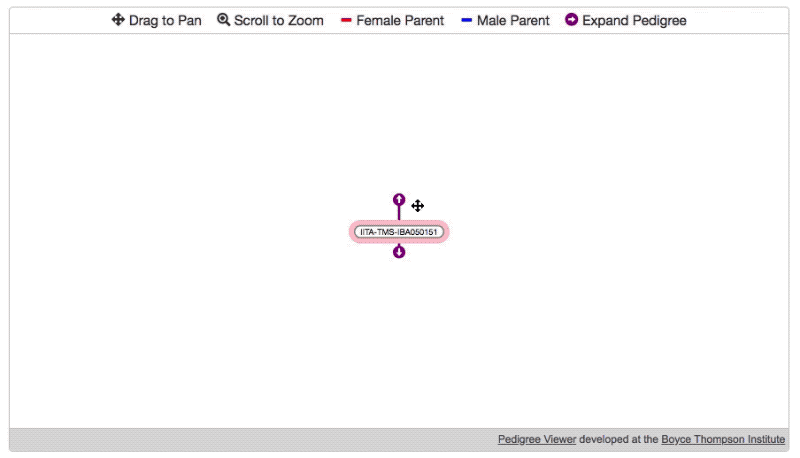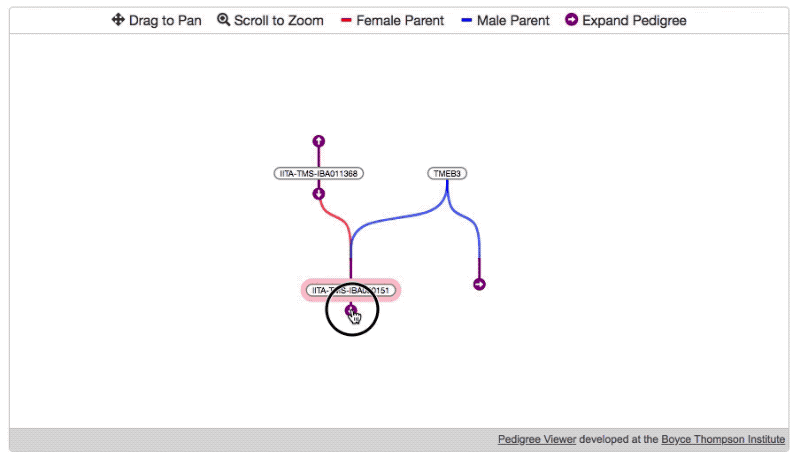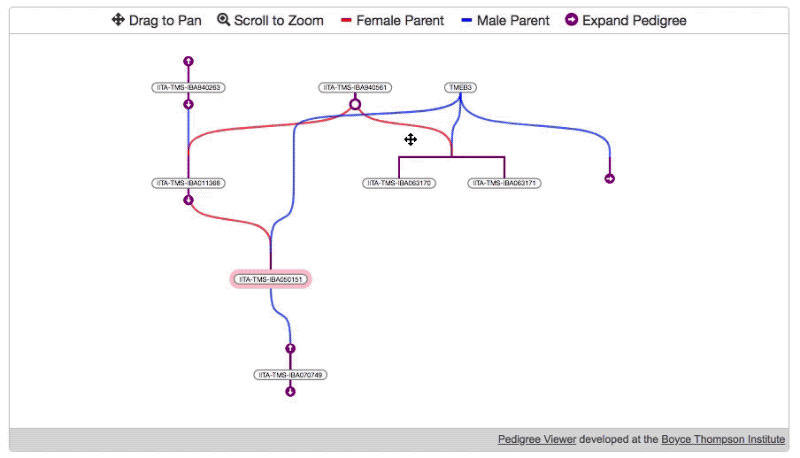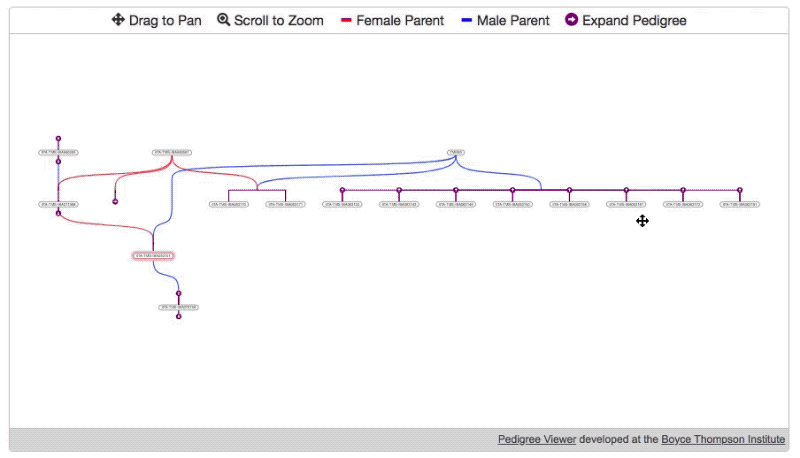
- Then, specify the root node and load the initial data.
pdg.newTree( //REQUIRED root germplasmDbId of germplasm which should be displayed on start (highlighted in pink) "1234" )
- Last, tell the viewer where to draw.
pdg.drawViewer( //REQUIRED selector for the parent node of the new viewer SVG element. "div#pdgv-wrap", //OPTIONAL width of viewer 800, //OPTIONAL height of viewer 400 );
- (Optional) Provided function to retrieve phenotype/marker data to display. (See the next section.)
Use the pdg.data(accessor) method to display marker or phenotype data. Data is displayed as a table to the right of each node in the tree. accessor should be a function which takes a germplasmDbId returns an array or Promise which will resolve to an array in the following format:
// Example of data accessor return value
[
{
"name":"DS", // Unique name for data row
"values":[ // Array of data values to display in row
{
"value":"0", // Text to display as data value
"color":"#EEE" // background color of data value
},...
]
},...
]// Source example
pdg.data(function(germplasmDbId){
return [
{name:"germplasmDbId",values:[
{value:germplasmDbId,color:"#ffb3ba"}
]}
]
}) // Source example
pdg.data(function(germplasmDbId){
return fetch(`https://mysite.com/api/marker_data/${germplasmDbId}`)
.then(resp=>resp.json())
.then(data=>data.markers);
/*
data.markers: [{
"name": "Marker:4:12202:REF",
"values": [{
"value": "REF",
"color": "#eeeeee"
}]
}, {
"name": "Marker:3:04993:GT",
"values": [{
"value": "T",
"color": "#baffc9"
}, {
"value": "A",
"color": "#ffb3ba"
}]
}, {
"name": "Marker:3:05002:GT",
"values": [{
"value": "A",
"color": "#ffb3ba"
}]
}]
*/
}) - @solgenomics/brapijs (Peer Requirement)
- D3.js (v4) (Peer Requirement, expected to be in scope)
- d3-pedigree-tree
- @solgenomics/brapp-wrapper (For building the example.)
- jQuery (v3) (For the forms in the example.)