Developed by Emine Topcu @ Bui Lab, University of Ottawa, 2023
in Silico Fish is an open-source software to model spinal control of motor movement in larval zebrafish. It is inspired by the code written by (Roussel et al. 2021), which is available on https://raw.githubusercontent.com/Bui-lab/Code. The software allows custom model generation.
The executable file can be downloaded from the Releases link.
(The explanations below are for version 2.0. Version 2.6 offers many other features that will be helpful to the researcher. If you intend to use the software, feel free to reach out to me through GitGub, or my personal Twitter account @EmineTheStudent. I would be happy to give you a quick tour so you can make the most of the tool.)
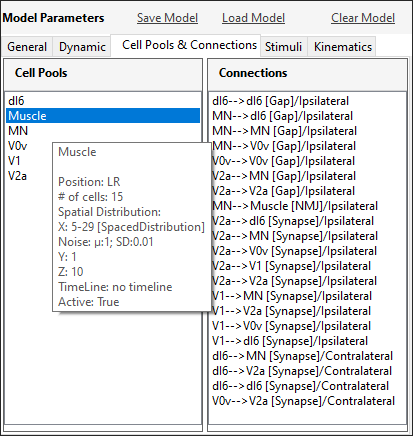
SiliFish allows you to define cell pools, connections between cell pools and stimuli.
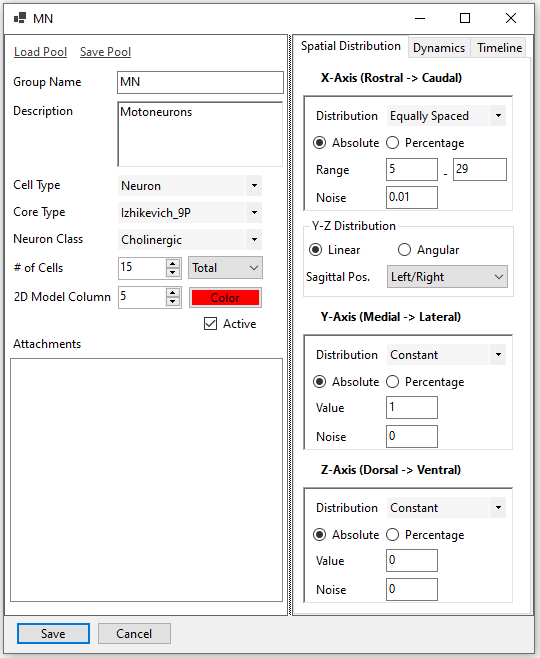
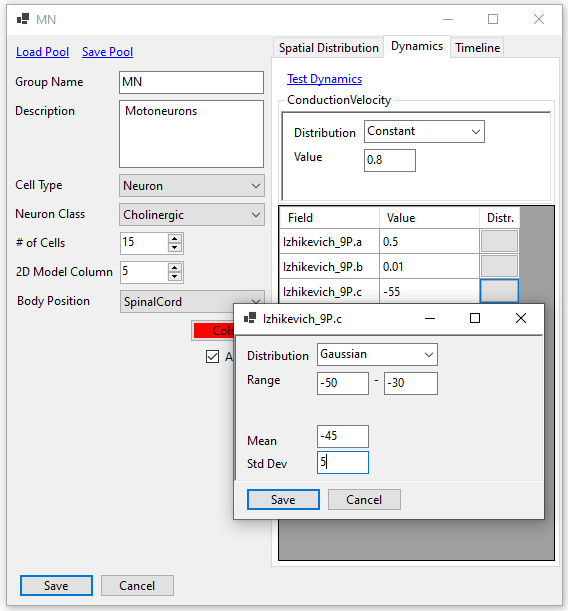
A cell pool consists of a specific number of cells. The cells can be spatially laid out in many ways by configuring the x, y, and z coordinates. Similarly, their intrinsic properties can be defined as distributions (Gaussian, uniform, etc.) rather than unique numbers. A timeline can also be defined to limit the active times of the cells in a cell pool.
Within the current version of SiliFish, there are various mathematical models that can be selected to model each cell group. The recommended one is Izhikevich-9P model, based on Izhikevich Simple Cells (Dynamical Systems in Neuroscience, p. 272).
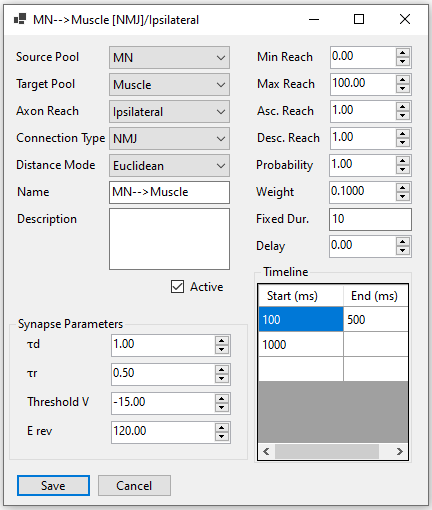
It is possible to define gap junctions or chemical synapses between two neurons or neuromuscular junctions between neurons and muscle cells. The connections are defined as probabilistic projections from one cell pool to another.
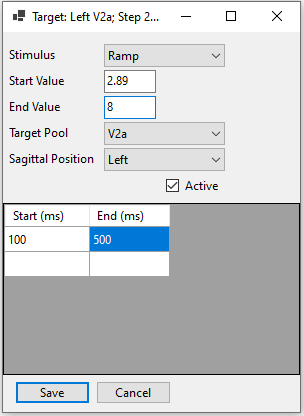
External stimuli applied to a specific cell pool can be defined.
SiliFish offers a visual tool to facilitate parameter fitting of the mathematical cellular model selected.
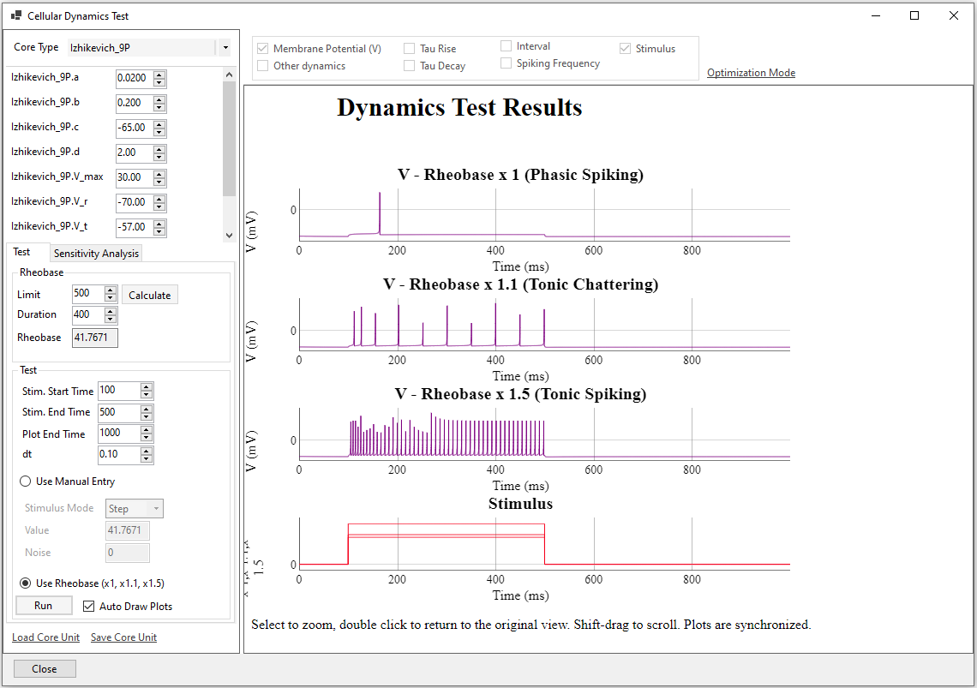
SiliFish incorporates a genetic algorithm facilitate parameter fitting of the mathematical cellular model selected.
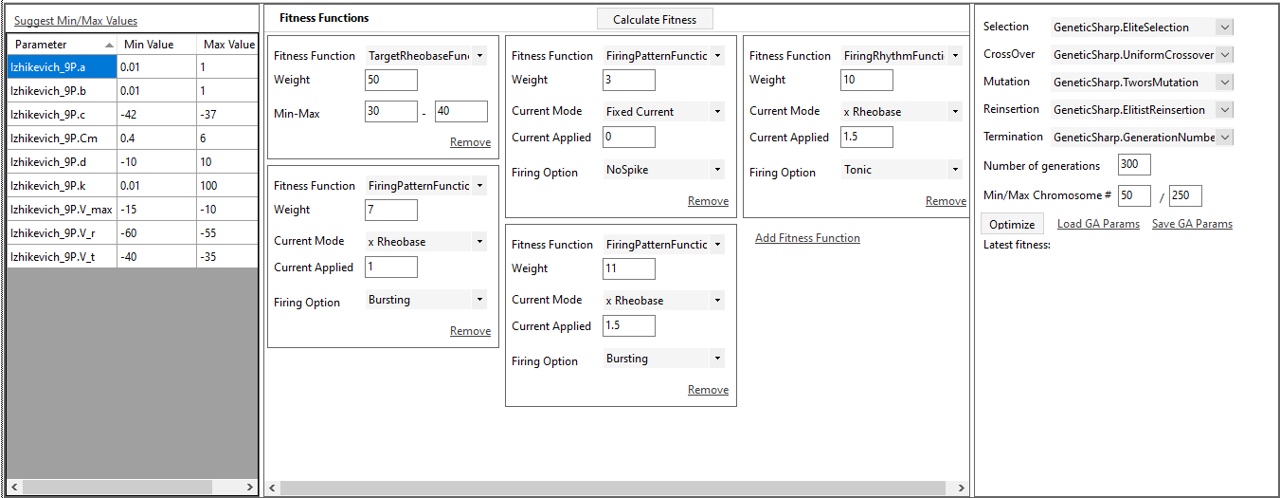
(The genetic algorithm uses GeneticSharp library (https://github.com/giacomelli/GeneticSharp) Please refer to https://github.com/Bui-lab/SiliFish/blob/main/3rd%20PARTY%20LICENSE for the licence information.)
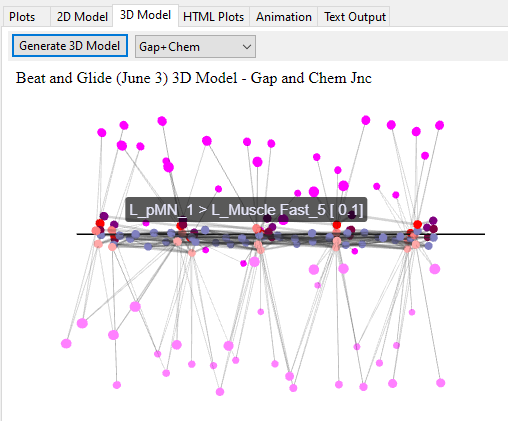
It is possible to check the accuracy of the model generated visually by the 2D and 3D outputs of the generated model.
(2D and 3D renderings use force-graph (https://github.com/vasturiano/force-graph) and 3d-force-graph (https://github.com/vasturiano/3d-force-graph) libraries. Please refer to https://github.com/Bui-lab/SiliFish/blob/main/3rd%20PARTY%20LICENSE for the licence information.)
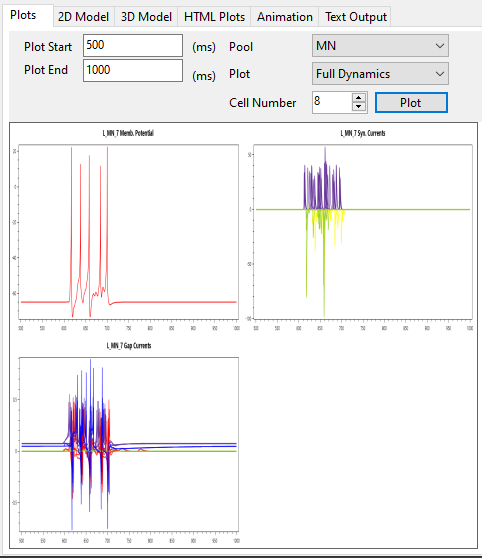
SiliFish has the capability to generate interactive and synchronous membrane potential, current, and stimulus plots for a single cell or group of cells.
(HTML based plots use dygraph (https://dygraphs.com/) libraries. Please refer to https://github.com/Bui-lab/SiliFish/blob/9eace2ba84584dcb0c95e2c582fa8f8d8ab64b60/3rd%20PARTY%20LICENSE for the licence information.)
The software is made available without any warranties. It is still under development. If you encounter any problems using the software or would like to suggest a new feature, please create a new issue through the Issues link.