Every experiment is sacred
Every experiment is great
If an experiment is wasted
God gets quite irate
Sacred is a tool to help you configure, organize, log and reproduce experiments. It is designed to do all the tedious overhead work that you need to do around your actual experiment in order to:
- keep track of all the parameters of your experiment
- easily run your experiment for different settings
- save configurations for individual runs in a database
- reproduce your results
Sacred achieves this through the following main mechanisms:
- ConfigScopes A very convenient way of the local variables in a function to define the parameters your experiment uses.
- Config Injection: You can access all parameters of your configuration from every function. They are automatically injected by name.
- Command-line interface: You get a powerful command-line interface for each experiment that you can use to change parameters and run different variants.
- Observers: Sacred provides Observers that log all kinds of information about your experiment, its dependencies, the configuration you used, the machine it is run on, and of course the result. These can be saved to a MongoDB, for easy access later.
- Automatic seeding helps controlling the randomness in your experiments, such that the results remain reproducible.
| Script to train an SVM on the iris dataset | The same script as a Sacred experiment |
from numpy.random import permutation
from sklearn import svm, datasets
C = 1.0
gamma = 0.7
iris = datasets.load_iris()
perm = permutation(iris.target.size)
iris.data = iris.data[perm]
iris.target = iris.target[perm]
clf = svm.SVC(C, 'rbf', gamma=gamma)
clf.fit(iris.data[:90],
iris.target[:90])
print(clf.score(iris.data[90:],
iris.target[90:])) |
from numpy.random import permutation
from sklearn import svm, datasets
from sacred import Experiment
ex = Experiment('iris_rbf_svm')
@ex.config
def cfg():
C = 1.0
gamma = 0.7
@ex.automain
def run(C, gamma):
iris = datasets.load_iris()
per = permutation(iris.target.size)
iris.data = iris.data[per]
iris.target = iris.target[per]
clf = svm.SVC(C, 'rbf', gamma=gamma)
clf.fit(iris.data[:90],
iris.target[:90])
return clf.score(iris.data[90:],
iris.target[90:]) |
The documentation is hosted at ReadTheDocs.
You can directly install it from the Python Package Index with pip:
pip install sacred
Or if you want to do it manually you can checkout the current version from git and install it yourself:
You might want to also install the numpy and the pymongo packages. They are optional dependencies but they offer some cool features:
pip install numpy, pymongo
The tests for sacred use the py.test package. You can execute them by running py.test in the sacred directory like this:
py.test
There is also a config file for tox so you can automatically run the tests for various python versions like this:
tox
At this point there are two frontends to the database entries created by sacred (that I'm aware of). They are developed externally as separate projects.
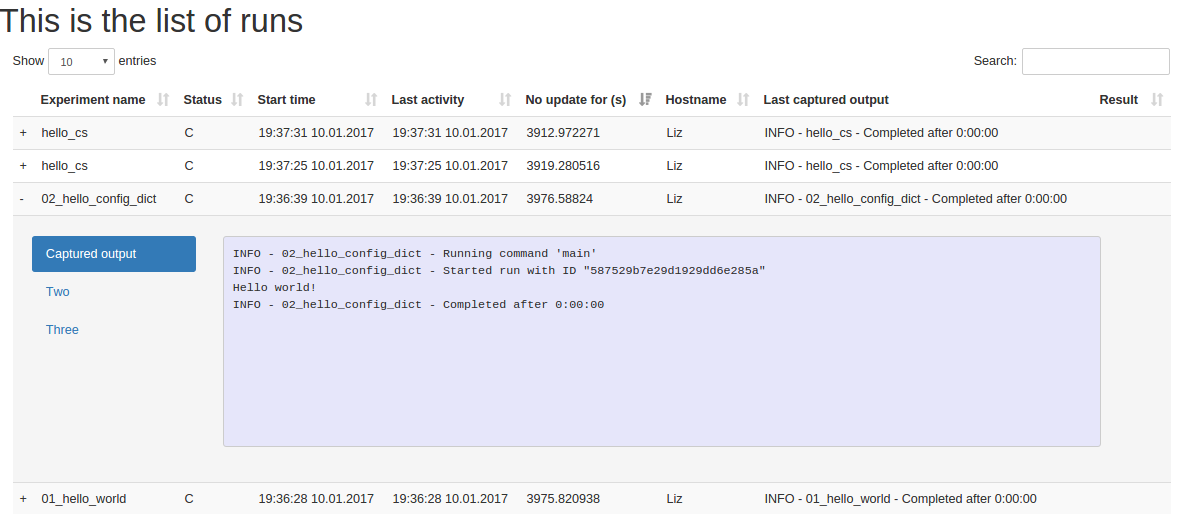
Sacredboard is a web-based dashboard interface to the sacred runs stored in a MongoDB.
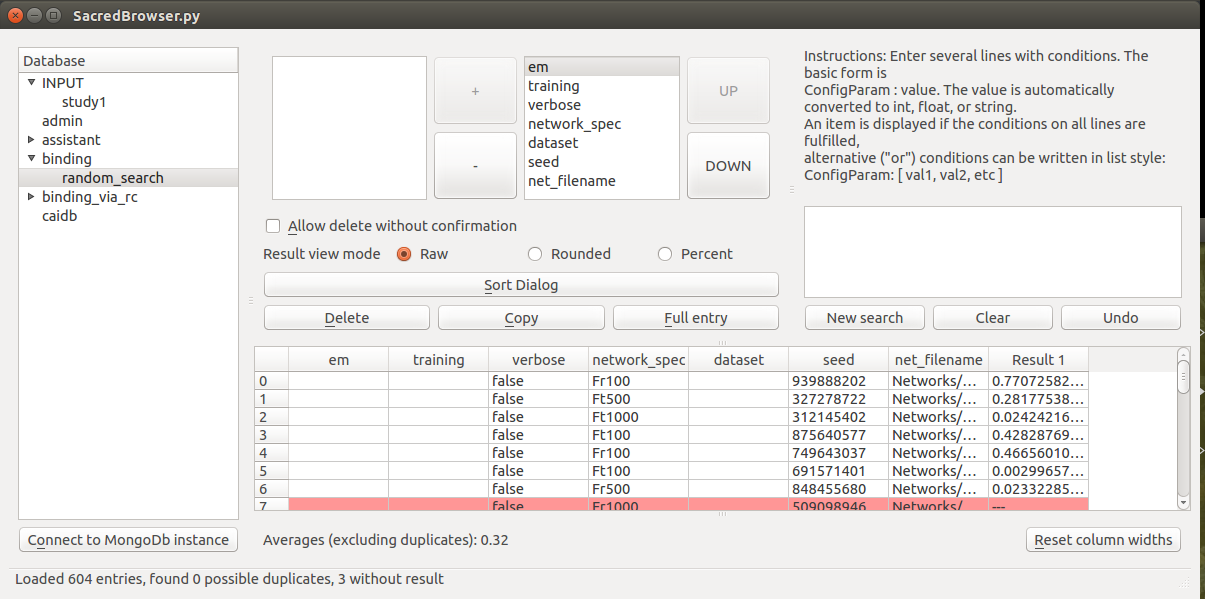
SacredBrowser is a PyQt4 application to browse the MongoDB entries created by sacred experiments. Features include custom queries, sorting of the results, access to the stored source-code, and many more. No installation is required and it can connect to a local database or over the network.
Prophet is an early prototype of a webinterface to the MongoDB entries created by sacred experiments, that is discontinued. It requires you to run RestHeart to access the database.
Sumatra is a tool for managing and tracking projects based on numerical
simulation and/or analysis, with the aim of supporting reproducible research.
It can be thought of as an automated electronic lab notebook for
computational projects.
Sumatra takes a different approach by providing commandline tools to initialize a project and then run arbitrary code (not just python). It tracks information about all runs in a SQL database and even provides a nice browser tool. It integrates less tightly with the code to be run, which makes it easily applicable to non-python experiments. But that also means it requires more setup for each experiment and configuration needs to be done using files. Use this project if you need to run non-python experiments, or are ok with the additional setup/configuration overhead.
FGLab is a machine learning dashboard, designed to make prototyping
experiments easier. Experiment details and results are sent to a database,
which allows analytics to be performed after their completion. The server
is FGLab, and the clients are FGMachines.
Similar to Sumatra, FGLab is an external tool that can keep track of runs from any program. Projects are configured via a JSON schema and the program needs to accept these configurations via command-line options. FGLab also takes the role of a basic scheduler by distributing runs over several machines.
By tracing system calls during program execution CDE creates a snapshot of all used files and libraries to guarantee the ability to reproduce any unix program execution. It only solves reproducibility, but it does so thoroughly.
This project is released under the terms of the MIT license.