A collection of Python files / libraries to implement the Postnova, Abeysuriya, Tekieh, et al. models of human photobiology from measured spectrophotometer data / CIE S 026 metrics / or multi-spectral simulations using ALFA. These models are based on the following papers:
- Postnova, S., Lockley, S. W., & Robinson, P. A. (2018). Prediction of cognitive performance and subjective sleepiness using a model of arousal dynamics. Journal of biological rhythms, 33(2), 203-218.
- Abeysuriya, R. G., Lockley, S. W., Robinson, P. A., & Postnova, S. (2018). A unified model of melatonin, 6-sulfatoxymelatonin, and sleep dynamics. Journal of Pineal Research, 64(4), e12474.
- Tekieh, T., Lockley, S. W., Robinson, P. A., McCloskey, S., Zobaer, M. S., & Postnova, S. (2020). Modeling melanopsin-mediated effects of light on circadian phase, melatonin suppression, and subjective sleepiness. Journal of pineal research, 69(3), e12681.
The code is licensed under GPL v3. For use of the code in research, kindly cite our associated publications from Building Simulation 2021:
- Jakubiec, J. A., & Alight, A. (2021). Spectral and biological simulation methods for the design of healthy circadian lighting. In Proceedings of Building Simulation 2021: Bruges.
- Alight, A., & Jakubiec, J. A. (2021). Evaluating the use of photobiology-driven alertness and health measures for circadian lighting design. In Proceedings of Building Simulation 2021: Bruges.
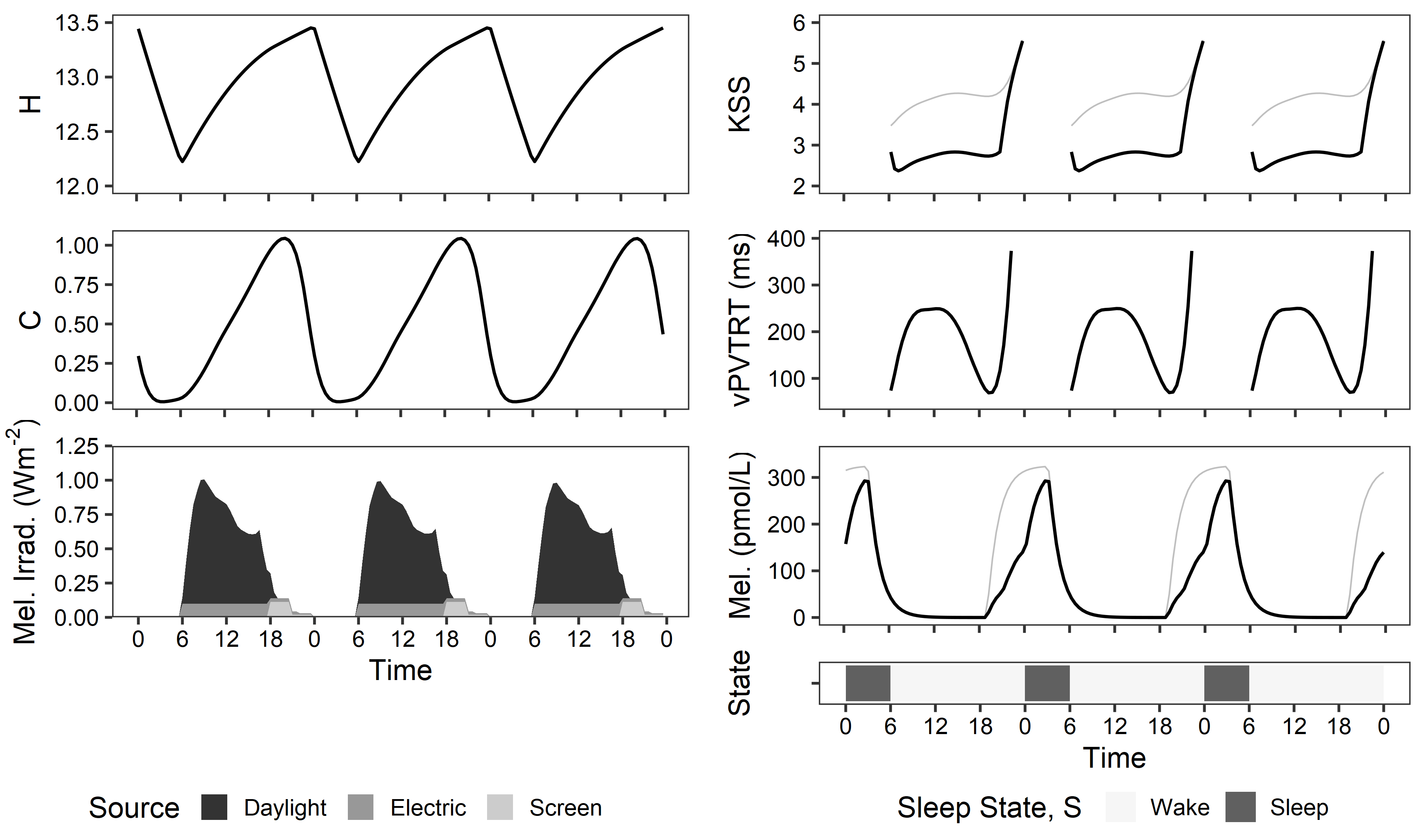
biological_model.pyandexample -files: Calculate the Postnova, Abeysuriya, and Tekieh, et al. models of photobiological dynamics based on sleep/wake dynamics and received light. Outputs include: subjective sleepiness (KSS), melatonin blood level concentration, reaction time (vPVTRT), and phase shifting.spectral_conversion.pyandexample -files: Convert ALFA spectral irradiance simulations to CIE S 026 outputs automatically.example - getting started.py: Perform simple photobiological modeling on a short timescale.example - annual lightsolve simulation run.pyandexample - process annual results.py: Perform annual photobiological modelling and analyze results seasonally.
All code is provided in the root folder.
*.pyfiles identified above include libraries and example use cases.*.csvfiles contain important response curve data for CIE S 026 (action_spectra.csv,D65.csv), EML (c-lambda_lucas2014), and photopic illuminance (v_lambda_1924.csv).- The two
example -folders contain ALFA-simulated results used in the examples.
- Python
>= 2.7 - Numpy
- Python 'Statistics' library
- Download the code using
git clone https://github.com/C38C/NIF_Photobiology.gitor directly from Github. - Explore the example files.