This repository provides access to a set of pipelines developed by CCBR members to analyze NGS data on DNAnexus. They allow a user to process raw data starting from FastQ files to reach common endpoints for downstream analysis such as a list of annotated mutations, a set of annotated peaks, or a raw counts matrix for differential expression analysis.
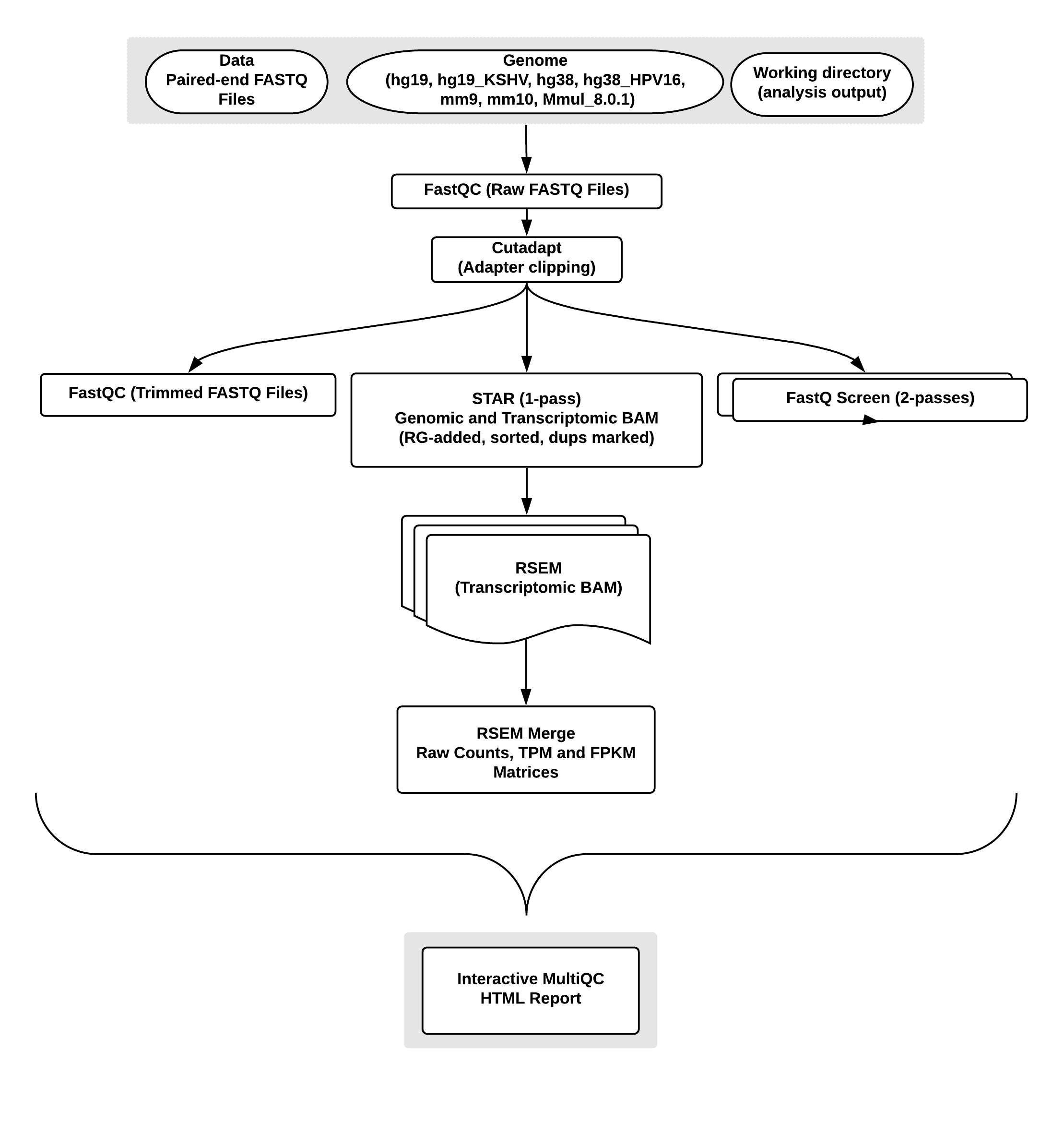
Overview of the RNA-seq pipeline:
FastQC: a quality control tool for high throughput sequence data, written by Simon Andrews at the Babraham Institute in Cambridge. It provides a modular set of analyses which can be used to give a quick impression of whether your data has any problems of which you should be aware before doing any further analysis.Cutadapt: a tool to find and remove adapter sequences, primers, poly-A tails and other types of unwanted sequence from your high-throughput sequencing reads. Cutadapt is being used to remove adapter sequences.FastQ Screen: a tool to screen a library of sequences against a set of sequence databases so one can see if the composition of the library matches with what is expected.STAR: an ultrafast splice-aware RNA-seq aligner.RSEM: quantifies gene and isoform expression levels from RNA-Seq data.MultiQC: aggregate results from bioinformatics analyses across many samples into a single report
RNA-seq Workflow
Center for Cancer Research
Collaborative Bioinformatics Resource
usage: dx run ccbr_RNA-seq_WF [-iARG_NAME=VALUE ...]
Inputs:
Input Fastq File Array: -iFastqFiles=(file) [-iFastqFiles=... [...]]
Please provide a series of paired-end FastQ files:
-iFastqFiles=sample1.R1.fastq.gz -iFastqFiles=sample1.R2.fastq.gz
-iFastqFiles=sample2.R1.fastq.gz -iFastqFiles=sample2.R2.fastq.gz
Reference Files for each Genome: -iReferenceFiles=(file)
Genomic Resource file: 'genome2resources.tsv':
See Example (file-Fjvv3100xx3f7J4p7fKFz907)
Version of STAR to use for alignment: '2.6.0a' or '2.7.0f' [default]: [-iStarVersion=(string, default="2.7.0f")]
Choices: 2.6.0a, 2.7.0f
Version of STAR to Run: '2.6.0a' or '2.7.0f'. Optional Argument.
Default is set to latest supported verison of STAR: '2.7.0f'.
Reference Genome: -iRefGenome=(string)
Reference Genome: Needed for dynamically resolving the correct reference files.
Choices: [mm9, mm10, mm10_M18,
hg38, hg38_v28, hg19,
hg38_HPV16, hg19_KSHV,
hs38d1, hs37d5, Mmul_8.0.1]
Note: mm10 was built using M21 GENCODE annotations and hg38 and hg38_HPV16 were built using version 30 GENCODE annotations.
module load DNAnexus # or download dx-toolkit and add to $PATH
dx login # optional step (if you have not logged in the last 30 days)
# prompt will ask for Username and Password
# select this Project: 'CCBR_RNASeq_Pipeline'
# Data upload (optional, if data not on DNAnexus)
## Assumes data exists in present working directory
dx upload control_1.R1.fastq.gz --destination=/Testing/
dx upload control_1.R2.fastq.gz --destination=/Testing/
dx upload treatment_1.R1.fastq.gz --destination=/Testing/
dx upload treatment_1.R2.fastq.gz --destination=/Testing/
# Run the RNA-seq Pipeline
## Supports mm9, mm10 (M21), hg38 (v30), hg38_HPV16 (v30), hg19, hg19_KSHV, hs38d1, hs37d5, Mmul_8.0.1
echo "Y n" | dx run /Workflow/ccbr_RNA-seq_WF \
-iFastqFiles=/Testing/control_1.R1.fastq.gz \
-iFastqFiles=/Testing/control_1.R2.fastq.gz \
-iFastqFiles=/Testing/treatment_1.R1.fastq.gz \
-iFastqFiles=/Testing/treatment_1.R2.fastq.gz \
-iRefGenome=hg19 \
-iReferenceFiles=file-Fjvv3100xx3f7J4p7fKFz907 \
--destination=/Testing/Example/ \
--priority=highFor more detailed documentation about RNA-seq and the pipeline, please checkout our user documentation page.
Step 0.) Log into DNAnexus
module load DNAnexus # module is available from helix/biowulf
dx login # optional step (if you have not logged in the last 30 days)
# prompt will ask for Username and PasswordStep 1.) Upload data to DNAnexus
# Assumes your FastQ files exists in your present working directory on biowulf/helix
# In this example, we will be uploading the data to a directory called /Testing/tmp/ on DNAnexus
# The directory must exist on DNAnexus prior to uploading data
# Here is the command to create a new directory:
dx mkdir /Testing/tmp/
# Upload files
dx upload input_rep1.R1.fastq.gz --destination=/Testing/tmp/
dx upload treatment_rep1.R1.fastq.gz --destination=/Testing/tmp/
dx upload input_rep2.R1.fastq.gz --destination=/Testing/tmp/
dx upload treatment_rep2.R1.fastq.gz --destination=/Testing/tmp/
# Optional: If you want to take a peek at the data you just uploaded, you could run something like this:
dx ls -la /Testing/tmp/
# To see all the dx commands available and get more info about them run:
dx help allAt the current moment, there are two ChIP-seq workflows on DNAnexus:
ccbr_chipseq_pipeline_single_end_two_replicatesccbr_chipseq_pipeline_single_end_no_replicates
In this example, we will walk through how to run the ccbr_chipseq_pipeline_single_end_two_replicates workflow, but keep in mind, the process is very similar for the no replicates workflow.
Note:
Both of these workflows support the following reference genomes: mm9, mm10, hg19, hg38.
The –-destination argument is the pipeline’s output directory on DNAnexus (if this directory does not exist, it will created it). Please make sure you filenames end with “.R1.fastq.gz” ~ similar to Pipeliner.
To get more information about any pipeline, you can run the following: dx run pipelinename -h
#Example:
dx run ccbr_chipseq_pipeline_single_end_two_replicates -hStep 2.) Run the ccbr_chipseq_pipeline_single_end_two_replicates pipeline on DNAnexus
## Pipeline supports mm9, mm10, hg38, hg19
echo "Y n" | dx run ccbr_chipseq_pipeline_single_end_two_replicates \
-iInputFastqRep1=/Testing/tmp/input_rep1.R1.fastq.gz \
-iTreatmentFastqRep1=/Testing/tmp/treatment_rep1.R1.fastq.gz \
-iInputFastqRep2=/Testing/tmp/input_rep2.R1.fastq.gz \
-iTreatmentFastqRep2=/Testing/tmp/treatment_rep2.R1.fastq.gz \
-iGenome=mm9 \
-iGenome2Resources= file-FXbpk100vbJF4qX02FV3F6K2 \
--destination=/Testing/tmp/Test_SE_2R_mm9/ \
--priority=highUnder Constuction: Coming Soon!