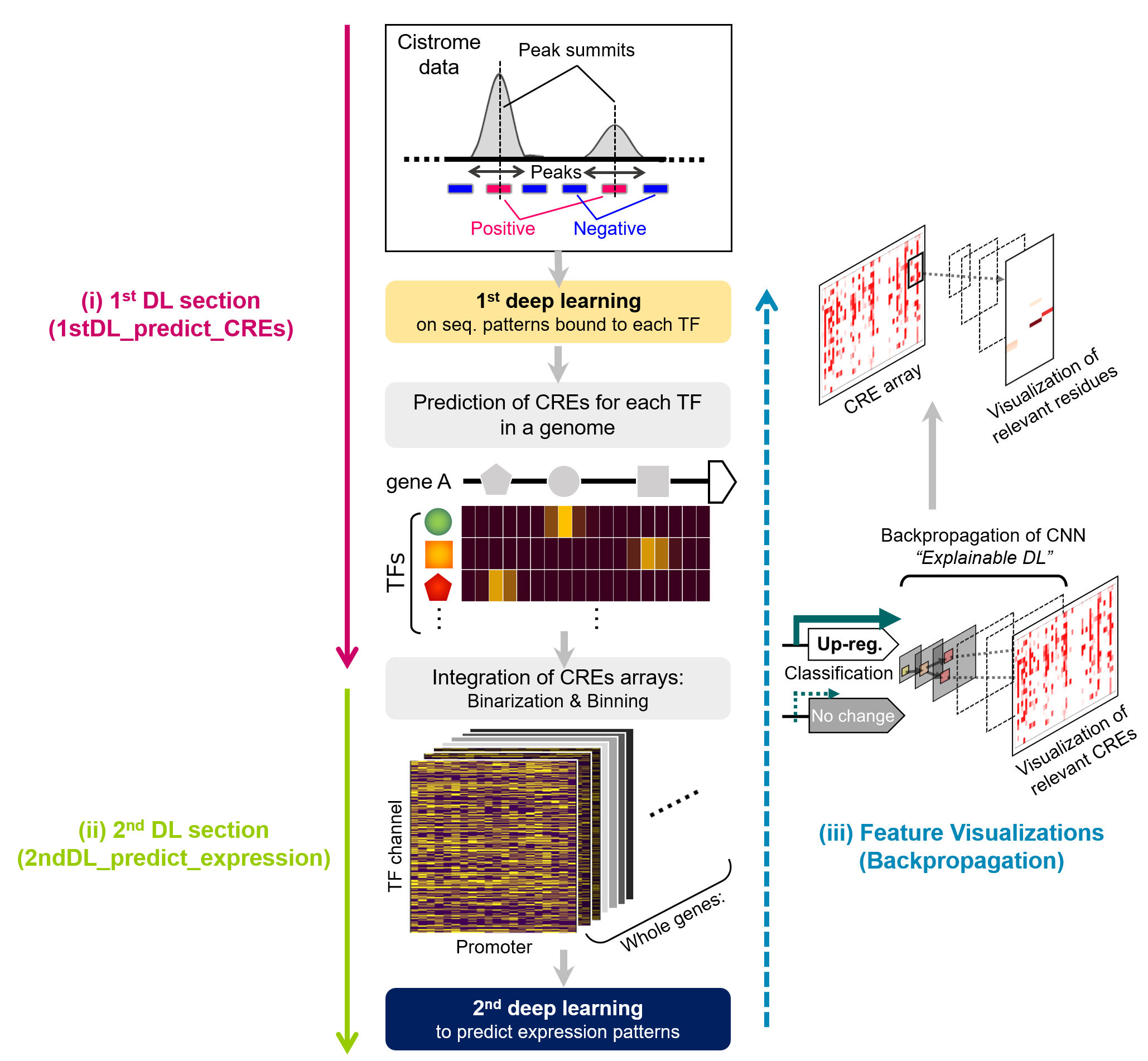
(i). Training of TFs recognition patterns from cistrome datasets [on directory “1stDL_predict_CREs”]
(iii). Feature visualization [on directory “Backpropagation”])
-
Extract fragments including significant peak from DAP-Seq (NOTE: the length should be identical), to convert into simple text files. If you target 31-bp tiles, save as .txt like,
ATGCGTGCGTGCGTGGCTGCAATGTGCAAAT GGGTACTAGCTTGTATATAGCAAATATAGCA -
Training/validation by a fully-connected model
python FC-cistrome-training.py [-p] [-n] [-o] [-e] [-l] option: "-p", help="File path to positive DNAs" "-n", help="File path to negative DNAs" "-o", help="output prefix" "-e", help="epoch numbers" "-l", help="DNA length"→ Output “.h5 file” with ROC-AUC data.
-
Detection of prediction (of each TF biding) in the target promoter sequences in bin sliding-windows.
Target promoter sequences should be in fasta format.
python MultiSeq_CREs_prediction_walking.py [-f] [-m] [-w] [-b] [-o] option: "-f", help="File path to fasta" "-m", help="File path to HDF5 or H5 model" "-w", help="walk bp size" "-b", help="bin bp size" (should be same as the tile set above) "-o", help="output file name"→ Output “wOTU_XXX(out)” with each target info as OTUs.
-
Conversion to binary CRE arrays in bins
python BinIntg2BinaryArray.py [-i] [-b] [-t] [-o] option: "-i", help="File path to input" "-b", help="bin size" "-t", help="confidence threshold (0-1)" "-o", help="output file name"With 2-bp walking size in the previous step (MultiSeq_CREs_prediction_walking.py), “-b 25” produces an array with 50-bp bins.
If input file is like
geneA 0.1 0.4 0.9 0.1 0.1 0.0 0.2 1.0 0.9-b 3 -t 0.8 produces
geneA 1 0 1
- Use “2ndDL_predict_expression” directory.
-
Move all of the data including transitions of the predicted TF-binding sites for all target promoters, into “raw_data”.
-
python make_dataset.py (--raw_data_root [directory including data])→ Output compiled data (train_00/, train_01/…) into /gene_dataset
-
Make target binary expression pattern” (with the identical OTU names).
target binary expression pattern file (with a specific name) is like (tab-delimited),
geneA 0 geneB 1 geneC 0 geneD 0 geneE 1Now, the file/code structure is like,
/root ├ 1dCNN_CisDecoding_training_basic.py ├ data_utils/generator.py ├ make_dataset.py ├ gene_dataset/ │ ├ train_00/ │ │ ├ >XXX.npy │ │ ├ >YYY.npy │ │ ⋮ │ │ └ >ZZZ.npy │ ├ train_01/ │ ├ train_02/ │ ⋮ │ └ train_09/ │ ├ binary_expr_pattern file (a specific name) ├ cnn_models/cnn_model_bisic.py -
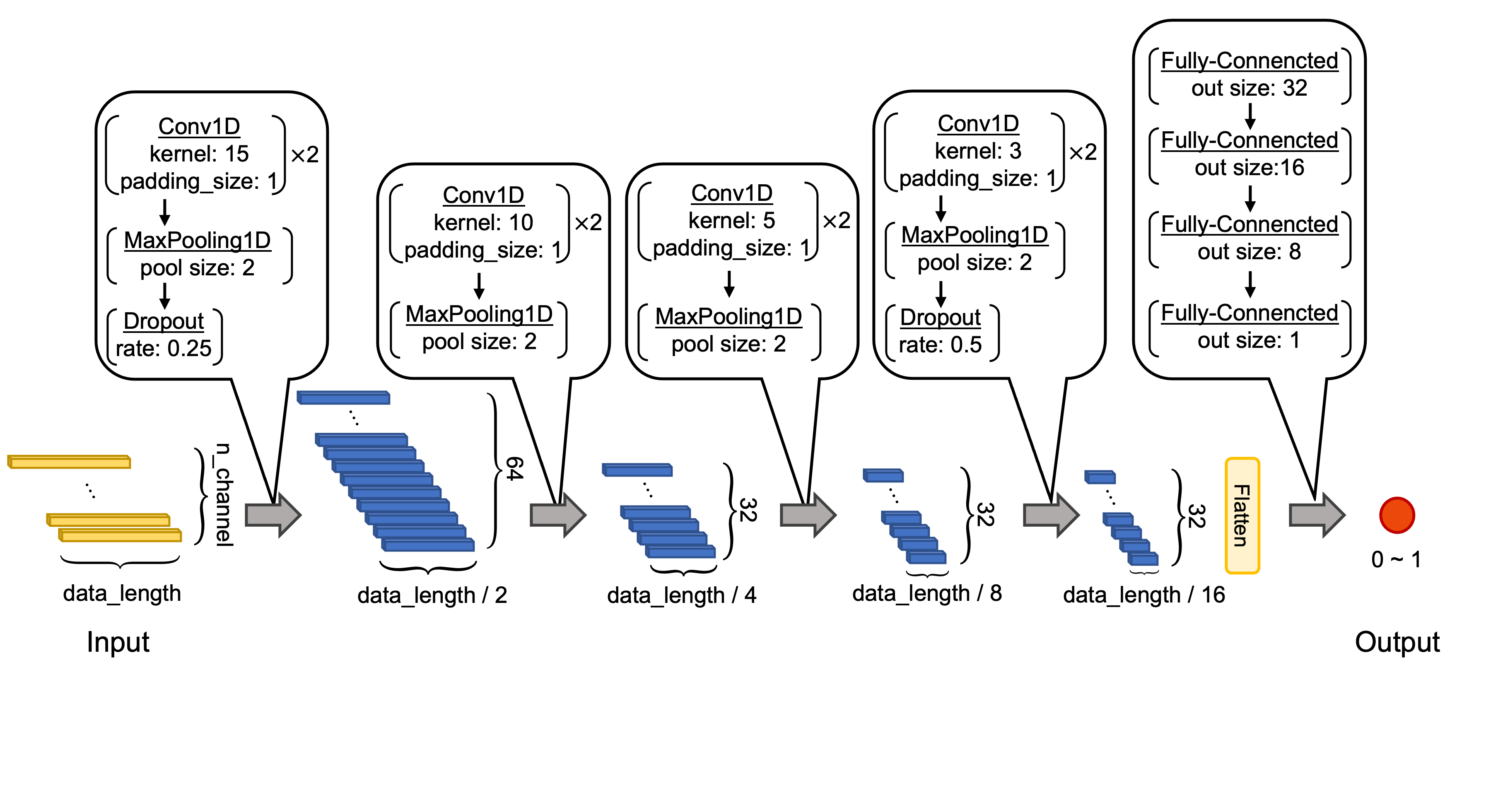
python 1dCNN_CisDecoding_training_basic.py [--n_channel] [--data_length] [--batch_size] [--epochs] [--val_rate] [--shuffle] [--class_weight] [--target_file] [--learning_rate] [--out_file] [--prediction_file]option: --n_channel', default=50, help='number of channels.' --data_length', default=20, help='length of sequence.' --batch_size', default=156, help='batch size for training.' --epochs', default=10, help='number of epochs for training.' --val_rate', default=0.3, help='rate of validation data.' --shuffle', default=True, help='phenotype data training shuffle' --class_weight', default=5, help='class-weight or positive sample imbalance rate' --target_file', default='BRup.txt', help='phenotype data file' --learning_rate', default=0.0001, help='learning rate' --out_file', default='model.h5', help='output model file name' --prediction_file', default='prediction.txt', help='output prediction confidence file name'→ Output trained h5 file, list for prediction confidence in validation datasets, ROC-AUC value and curve, and confusion matrix.
- Use “Backpropagation” directory. This step requires “jupyter notebook”, handling “ipynb” format
-
GuidedBackProp_CisDecode_batch.ipynb(need “visualizations_forCisDecode.py” and “helper_forCisDecode2.py” in the same directory.) Open jupyter, and run the ipynb file. -
NOTE: At the third cell given below, we may have to repeat runs of this cell until the “dense” name is properly changed (expect 4-times)
partial_model = Model( inputs=model.inputs, outputs=iutils.keras.graph.pre_softmax_tensors(model.outputs), name=model.name, ) partial_model.summary() -
Need the trained prediction model “XXX.h5”, and “YYY.npy” for the objective genes, which have been made in the “make_dataset.py” section in the section (ii) above.
-
The objective gene files (in npy format) need to be located on “select_GBP” directory.
-
GuidedBackprop_CREs-prediction.ipynb(need “visualizations_forCisDecode.py” and “helper_forCisDecode2” in the same directory.) -
Need a prediction model “ZZZ.h5” for each TF channel, which has been made in the section (i), and “fragments list” for the objective tiles, which would be made from a promoter sequence.