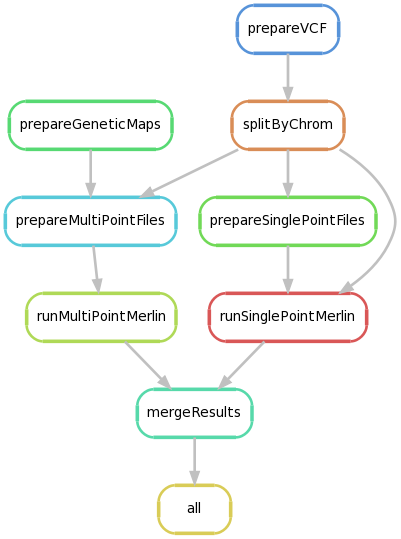
LodSeq performs the genetic linkage analysis across families, by computing lod-scores given a gvcf file and a related tfam pedigree file.
Both singlepoint and multipoint parametric linkage analyses are computed using merlin (Abecasis et al. Nature Genetics 2002).
Please cite LodSeq using the following DOI: 10.5281/zenodo.1321178.
Download genetic maps from the HapMap project:
$ cd data/inputs/
$ wget -nc ftp://ftp.ncbi.nlm.nih.gov/hapmap/recombination/2011-01_phaseII_B37/genetic_map_HapMapII_GRCh37.tar.gz
$ mkdir genetic_map_HapMapII_GRCh37 \
&& tar -zxf genetic_map_HapMapII_GRCh37.tar.gz -C genetic_map_HapMapII_GRCh37
$ cd ../..Once the prerequisites have been installed, configure the environment. vcftools, plink and merlin will be installed by conda:
$ conda env create -n lodseq --file environment.yamlThen, check that the external programs have been properly installed:
$ source activate lodseq
$ which vcftools && vcftools
$ which plink && plink
$ which merlin && merlin
$ source deactivate lodseqThe quick start dataset was arbitrarily modified from this available dataset:
https://github.com/jmchilton/SnpEffect/tree/master/papers/Protocol/data/chr7.vcf.gz
- The pedigree was reduced to 6 individus.
- Some genotypes were modified replacing phased genotypes by unphased genotypes.
- Some records were removed to reduce the final file size.
Activate the environment
$ source activate lodseqRun the workflow (using 2 threads)
$ mkdir test/
$ snakemake -s Snakefile --configfile config.yaml -j 2Exit
$ source deactivate lodseqThe lod-score results are output into directory test/.
You can choose a different output directory by changing the value of the field out_dir into config.yaml.
Please compare your results to the expected output files:
$ cd data/outputs/mergeResults/
$ ls -1
results_multipoint_merged_dominant.txt
results_multipoint_merged_recessive.txt
results_singlepoint_merged_dominant.txt
results_singlepoint_merged_recessive.txtRESULT EXAMPLE
Parametric singlepoint analysis using a recessive model:
$ less test/runSinglePointMerlin/7/results_singlepoint_chr7_recessive.txt
Parametric Analysis, Model Recessive_Model
=======================================================
POSITION LOD ALPHA HLOD
7:16487 0.000 0.000 0.000
7:16671 0.000 0.000 0.000
7:16692 0.000 0.000 0.000
7:16712 0.000 0.000 0.000
7:16717 0.000 0.000 0.000
7:16719 0.000 0.000 0.000
7:16787 0.000 0.000 0.000
7:16798 0.000 0.000 0.000
7:16878 0.000 0.000 0.000
7:17881 0.000 0.000 0.000
7:18510 0.000 0.000 0.000
7:19181 -2.575 0.000 0.000See documentation of merlin for a complete description.
WARNINGS
Loci with bad inheritance or loci with multichar allele variation will have a null lod-score (-0.000 or 0.000).
Edit the file config.yaml to change parameters of the workflow.
Description of the fields into config.yaml:
vcf # input gvcf file (.vcf or .vcf.gz)
tfam # input pedigree tfam file
dom_model # dominant model file
rec_model # recessive model file
genetic_maps # path of the HapMap genetic maps directory
out_dir # path of the directory containing LodSeq output files
out_log # path of a log file
out_prefix # prefix of output files
lod_threshold # minimal significant lod-score value, must be greater than 0
threads # number of threads used by multithread steps
chromosomes # list of chromosomes to analyzeYou can also change the parameters of the recessive and dominant models:
$ less data/inputs/parametric_dominant.model
$ less data/inputs/parametric_recessive.modelThe parameters of these models are respectively:
- affection name
- disease allele frequency
- penetrances
- model name
See a complete description and examples on merlin website and here.
Edith Le Floch, Centre National de Recherche en Génomique Humaine - CEA, Evry, France, edith.le-floch@cea.fr
Elise Larsonneur, Centre National de Recherche en Génomique Humaine - CEA, Evry, France, elise.larsonneur@cea.fr
For help please contact the authors.
LodSeq is released under the terms of the CeCILL license, a free software license agreement adapted to both international and French legal matters that is fully compatible with the GNU GPL, GNU Affero GPL and/or EUPL license.
For further details see LICENSE file or check out http://www.cecill.info/.