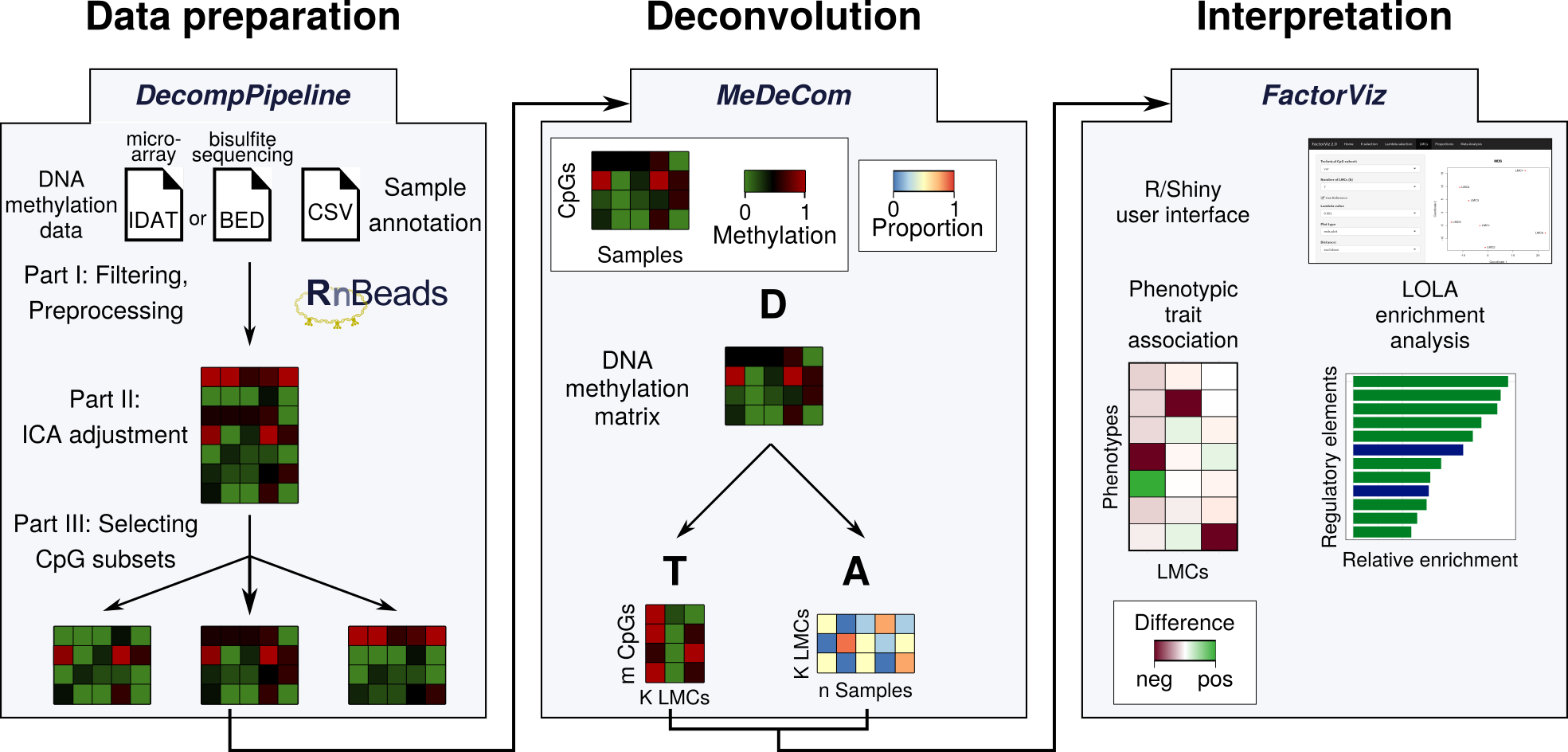
DecompPipeline provides a comprehensive list of preprocessing functions for performing reference-free deconvolution of complex DNA methylation data. It is an integral part of a recently published protocol for reference-free deconvolution and is independent of the deconvolution tool used.
DecompPipeline can be directly installed from GitHub within an R session on Linux systems. For macOS, we provide a binary version of MeDeCom, which needs to be installed prior to installing DecompPipeline. However, for Windows operating systems, a MeDeCom and thus DecompPipeline cannot be directly installed through R, but we provide a Docker image with all packages installed.
install.packages("devtools")
devtools::install_github("CompEpigen/DecompPipeline")
Installation has been tested on the following operating systems:
| Type | OS | Version | R-version | Installation successful | Protocol tested | Comments |
|---|---|---|---|---|---|---|
| Linux | Debian | 7 | R-3.5.2 | Yes | Yes | |
| Linux | Debian | 7 | R-3.6.0 | Yes | Yes | |
| Linux | Debian | 8 | R-3.5.3 | Yes | Yes (reduced) | |
| Linux | Debian | 8 | R-3.6.1 | Yes | No | |
| Linux | Debian | 8 | R-4.0 | Yes | No | |
| Linux | Debian | 10 | R-3.5.2 | Yes | Yes (reduced) | |
| Linux | Fedora | 28 | R-3.5.3 | Yes | Yes (reduced) | |
| Linux | Fedora | 31 | R-3.6.1 | Yes | Yes (reduced) | `igraph' dependency fails to install |
| Linux | CentOS | 8.0 | R-3.5.2 | Yes | Yes (reduced) | |
| Linux | CentOS | 8.0 | R-3.6.1 | Yes | Yes (reduced) | |
| Linux | Ubuntu | 19 | R-3.6.1 | Yes | Yes (reduced) | |
| MacOS | 10.14 | R-3.5.1 | Yes | Yes (reduced) | binary release used | |
| MacOS | 10.15 | R-3.6.0 | Yes | Yes (reduced) | ||
| Windows | 10 | Pro | R-3.6.1 | Yes | Yes (reduced) | Use the `windows' branch of MeDeCom |
| Docker | Debian | 10 | R-3.6.2 | Yes | Yes (reduced) | Docker image available |
| Docker | Windows | 10 | R-3.6.2 | Yes | Yes (reduced) | Docker image available |
In the reduced protocol, we executed preprocessing and a single MeDeCom run on a reduced dataset.
DecompPipeline includes three major steps, all of them are extensively documented. A more detailed introduction into DecompPipeline can be found in the package vignette and in the protocol .
There are dedicated preprocessing steps for both array-based data sets (prepare_data) and sequencing-based data sets (prepare_data_BS).
To select a subset of CpGs for downstream deconvolution analysis, the function prepare_CG_subsets can be used.
After these preprocessing steps, a deconvolution run can be started using DecompPipeline by envoking start_decomp_pipeline.
We also provide a wrapper functions that executes all the above steps in a single command (start_decomp_pipeline).
If you use DecompPipeline, please cite:
Scherer, M., Nazarov, P. V., Toth, R., Sahay, S., Kaoma, T., Maurer, V., Vedeneev, N., Plass, C., Lengauer, T., Walter, J., & Lutsik, P. (2020). Reference-free deconvolution, visualization and interpretation of complex DNA methylation data using DecompPipeline, MeDeCom and FactorViz. Nature Protocols, 15(10), 3240–3263. https://doi.org/10.1038/s41596-020-0369-6