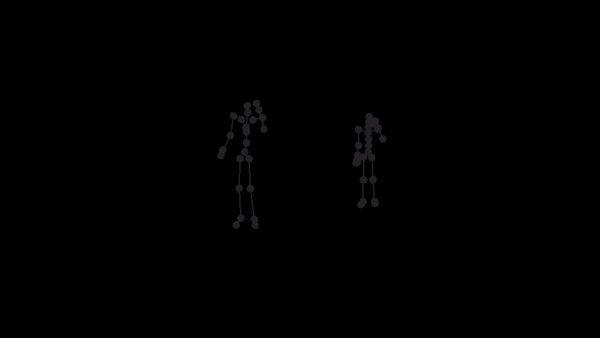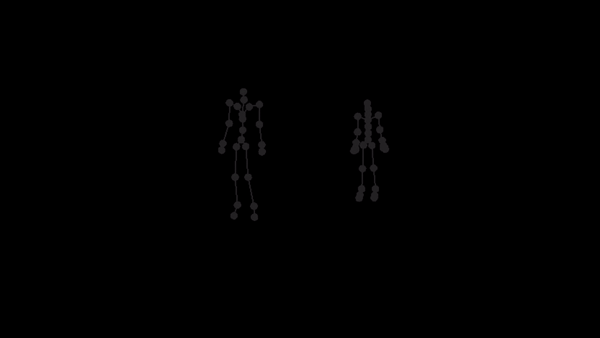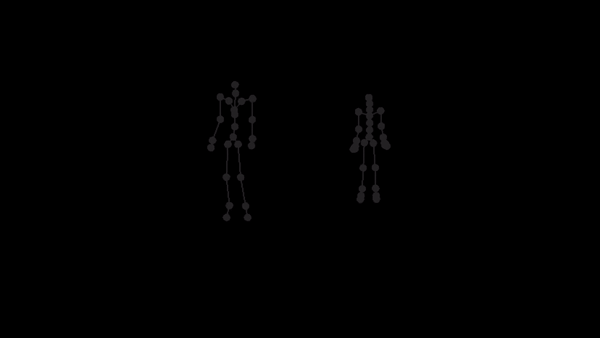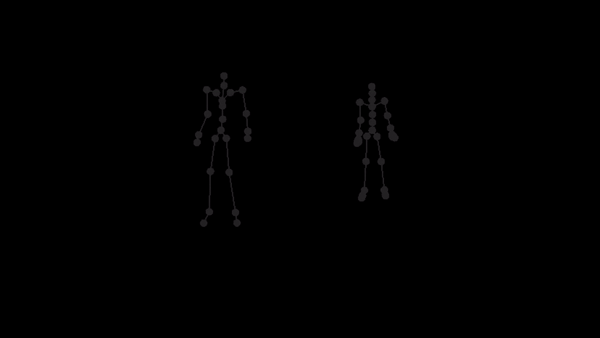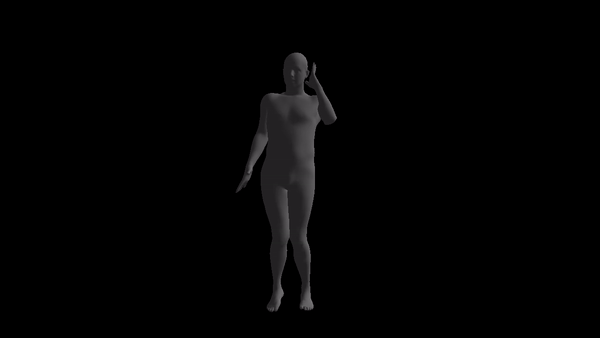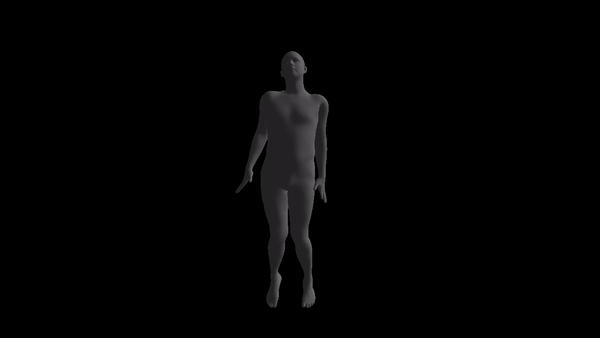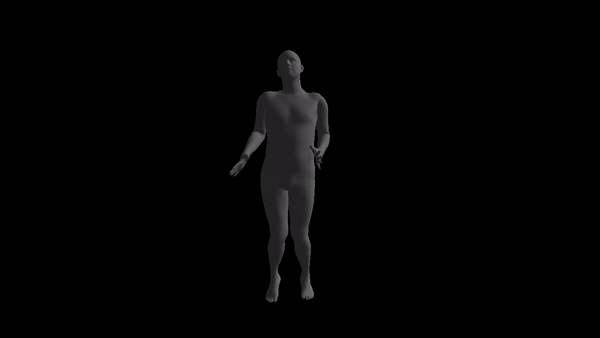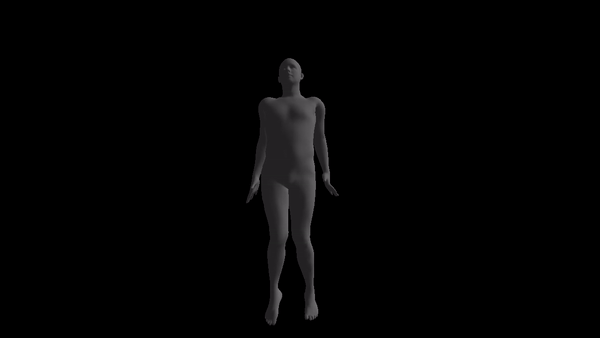NOTE: the code style is awful and I don't have time to prettify it. If anyone is really going to use this, please open an issue and I'll response as soon as possible.
For a given AMC/ASF motion sequence, we transfer the motion to SMPL model, and generate a corresponding 3D SMPL sequence.
This work is based on my implmentation of SMPL model and my implementation of AMC/ASF parser.
Run python 3Dviewer.py to see demo.
Also, run python batch.py to extract all poses into ./pose/ from ./data/.
-
Use
reader.parse_asf()to extract skeleton definition from.asffile:joints = reader.parse_asf(asf_path) -
Use
reader.parse_amc()to extract motion sequence from.amcfile:motions = reader.parse_amc(amc_path) -
Construct a
smpl_np.SMPLModelobject:smpl = SMPLModel(smpl_model_path) -
Construct a
imitator.Imitatorobject:imit = Imitator(joints, smpl) -
Use
imitator.Imitator.imitateto manipulate SMPL model to some pose:imit.imitate(motions[frame_index]) -
Use
smpl_np.output_meshto get.objfile:imit.smpl.output_mesh(output_path)
In step 5, the SMPLModel inside Imitator is set to the same pose as motions[frame_idx].
For any questions, feel free to open an issue.
The skeleton of SMPL is a little bit different from CMU MoCap Dataset's. In this implementation, we only process femur and tibia and ignore other differences. We first pose SMPL skeleton (specifically legs) to be in the same pose with ASF defination. After that, we extract rotation matrices from AMC files and apply them to the aligned SMPL model.
Feel free to contact me for more details.