In an effort to reduce the amount of strain on the Pennsylvania Department of Health websites during the peak hours (especially around noon when the daily updated information is posted), this script was created to quickly and easily parse the information without any additional overhead.
The script can optionally highlight certain counties which you may be interested in- in my case, I'm interested in Lancaster and Schuylkill counties. If you wish to highlight different counties, simply update the array in the script.
I gave a brief talk about this repository as part of TechLancaster. The video is available here.
To use this script, simply run it:
python covid.py
It's recommended that a virtualenv is used, for sanity reasons:
virtualenv env
source env/bin/activate
pip install -r requirements.txt
python covid.py
NOTE: Please be mindful that this script will still (in a minor way) affect the bandwidth of the host. Please do not use this script to poll the site more often than once every hour or so. The data does not change that often.
Ideally, the daily cases and fatalities would be stored in a database for ease
of use and to allow for powerful querying features.
In an effort to allow for easy duplication of the setup, a schema.sql file is
provided to allow for a duplicate Postgres database to be used.
To set this up, ensure you have a working Postgres database, then run
psql < schema.sql
You may then load the data as you see fit, including external sources as referenced in the schema file. The schema file is annotated with comments indicating each section and it's usage.
For convenience, a daily update SQL script has been provided as well, allowing for easy and quick updating of the various stats which are not provided by the Department of Health.
A further bonus of having the information in a database is the ability to hook external tools into it for visualization and data processing uses, such as Grafana.
The scripts and data used to generate the graphs are also included in the
repository.
The data is not automatically populated in to the gnuplot data file, it is
instead calculated separately following the daily PA Dept of Health update,
and provided as described in this repository, so that careful attention can be
provided to ensure validity.
Simply install gnuplot and run:
gnuplot plot_PA_Cases.gpi
The image file will be updated at cases.png
Typically, a daily update process is as follows:
- Insert the daily data into the db:
INSERT INTO covid19.covid19pa (date, confirmed, deaths) VALUES ("YYYY-MM-DD", ###, ###);
- Run the daily update script:
psql --file=daily-update.sql
- Plot the data:
gnuplot plot_PA_Cases.gpi
- Update any daily events, if necessary:
INSERT INTO covid19.pa_events (time, text) VALUES ('YYYY-MM-DD HH:MM:SS-4', 'Event');
Putting it all together, as a one-liner, this is my daily update script:
psql -d $db_name -h $hostname -c "insert into covid19.covid19pa \
(date, confirmed, deaths) values \
(CURRENT_DATE, 77999, 6162);"; \
psql -d $db_name -h $hostname --file=daily-update.sql; \
gnuplot plot_PA_Cases.gpi; \
git add .; \
git diff --staged; \
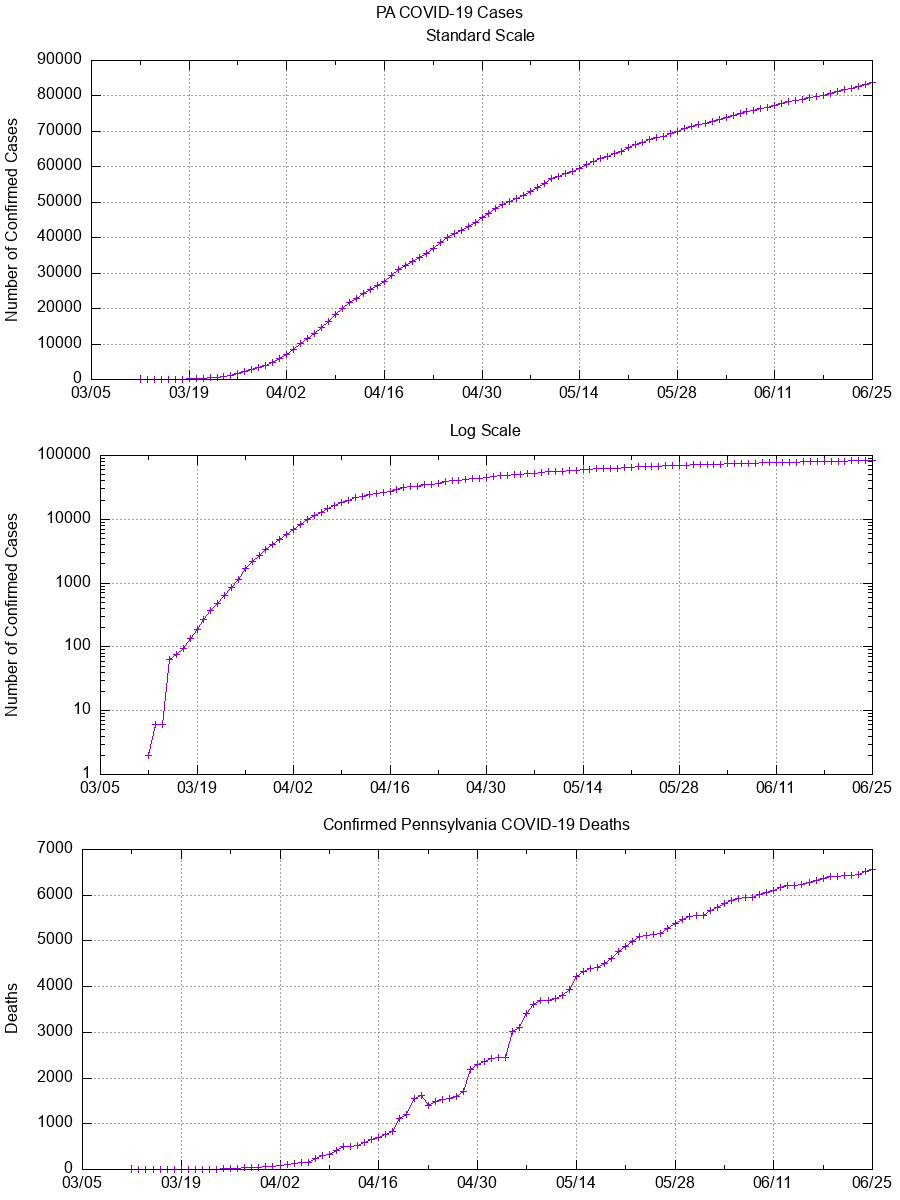
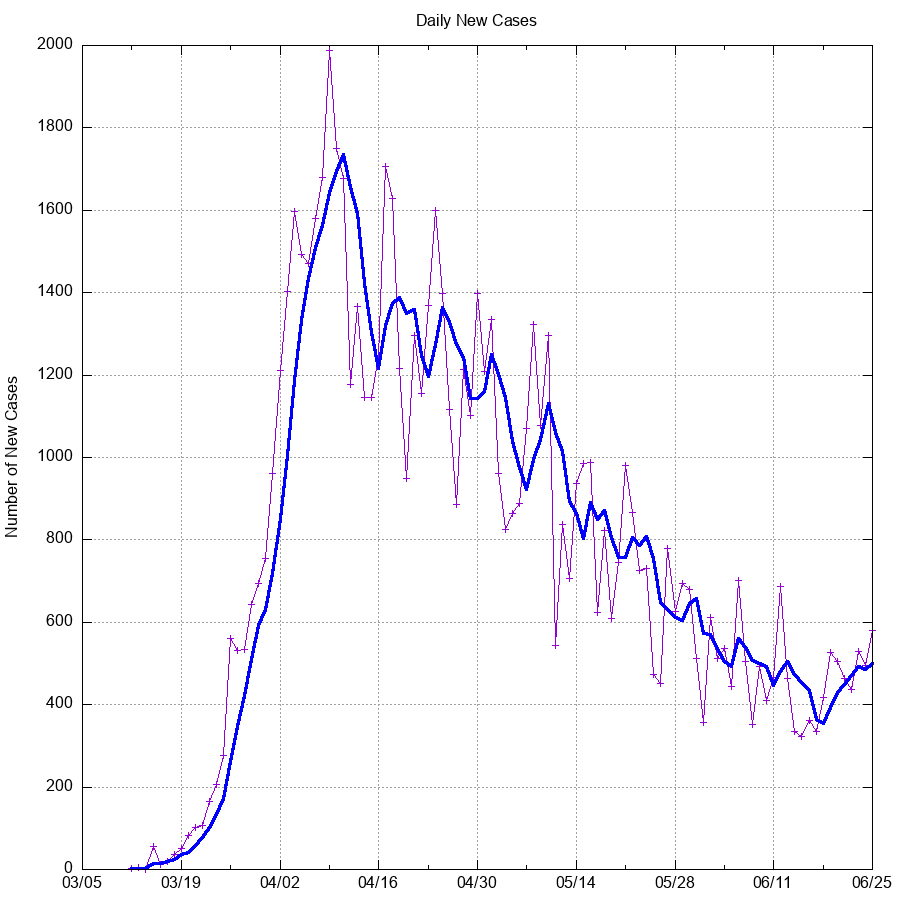
git commit -m "$(date +"%d %b %Y") Data";Below are graphs of confirmed cases, fatalities, and other potentially useful
data generated using the most recent available data and gnuplot.
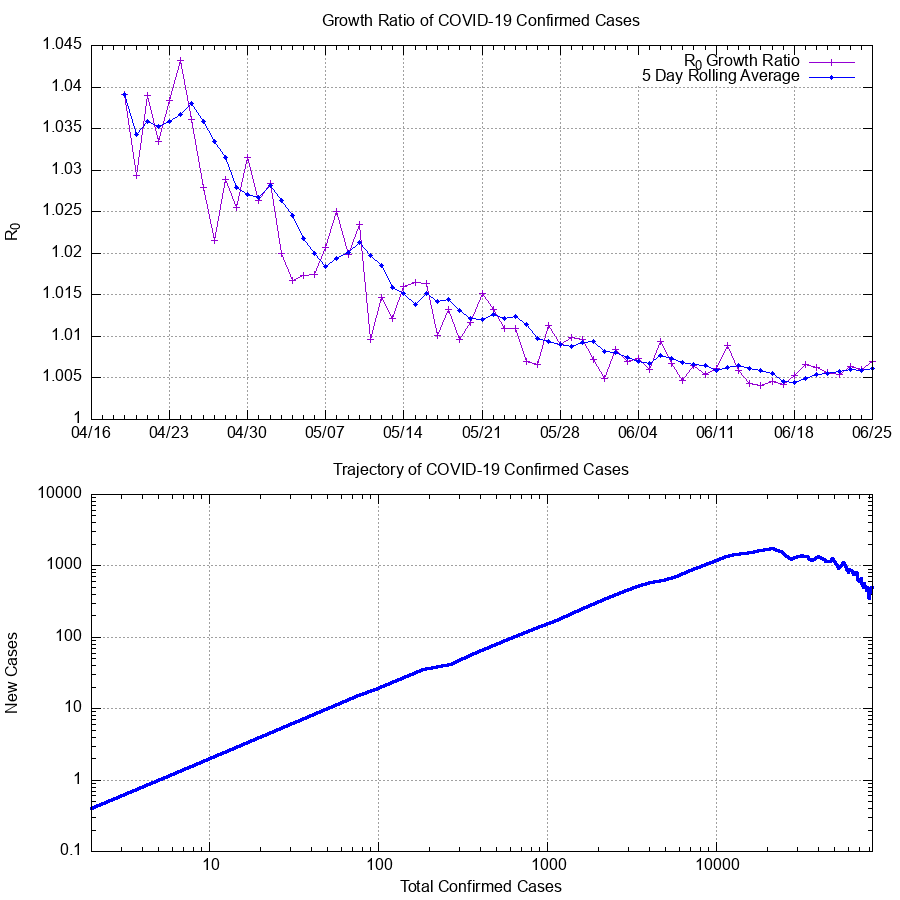
R0 is the 'Growth Factor'. In the most basic terms, this is how many new cases are created by existing cases.
For more information on how to read the trajectory graph, please see this excellent video by Henry Reich of MinutePhysics.
The only two datapoints provided by the PA Department of Health are
cases and deaths.
The other values are calculated using reasonable estimations and approximations
to the scientific models.
This section will describe how the calculations are performed and the formulae
for each value shown in cases.data.
Variables:
- Ct: Today's value for cases
- Ct-1: Yesterday's value for cases
- New Cases: The difference between the current value and the previous days value.
- Ct - Ct-1
- Growth Ratio: The ratio of current cases to previous days cases. Rough approximation of the "R0 Infectivity Rate".
- Ct / Ct-1
- Estimated Day: The estimated number of cases for the day, based upon the growth ratio.
- Ct-1 * Growth Ratiot-1
- Off By: The difference between the estimate and the actual daily count.
- -1 * (Estimated Dayt - Ct)
- Pct Error: The percent error calculated for the difference between the estimated count and the actual count.
- ABS((Estimated Dayt - Ct) / Ct)
Also of note, the daily-update.sql file includes the full calculations for
each of these items, allowing the database to calculate the values automatically.
As of 22 Apr 2020, the PA Dept of Health began providing 'probable' cases and deaths, alongside confirmed data. Because until this point, all data were assumed to be probable (and most likely) the calculations will continue to use this metric. Reasonable accommodations will be made to integrate the probable cases into error calculations, where possible. It's also important to note that the numbers do not identify which counties in particular are included in the 'probable' counts, so it's presently impossible do differentiate or reconcile the numbers.
This information is provided as part of an amateur attempt to analyze the data. This information should not be interpreted as exact scientific analysis, nor has it been peer reviewed in any way. Nonetheless, every attempt has been made to validate the accuracy of the numbers, such as providing a value of percent-error and a steadfast policy of +1 day estimations, only.
The only 'exact' values are those for the daily cases, deaths, and net daily-new cases, insofar as they have been reported by the Pennsylvania Department of Heath. For example, the values for R0 are approximations, at best. For more precise R0 calculations, other sources should be used instead.
For a more scientific and precise analysis, it is recommended that you seek out reporting performed by actual epidemiologists and data scientists.
-
27 Apr 2020: Updated method for creating the data file. Previously, data for each day was computed by manual processes in a spreadsheet. As of today, all calculated values are processed by the database and provided as output (i.e. there is no spreadsheet involved anymore). As a result, this allows for a more 'open' view of calculations, since the
.sqlfile used to update the daily values can also be provided for replication elsewhere. Notably, the plotted output does not change between this and the parent commit, strongly indicating that there are no changes to the fundamental data. -
7 May 2020: Updated plot method for Trajectory graph. This averaging plot (as opposed to the previous exact data plot) more closely matches the ideals and functions behind the original plot design from MinutePhysics. It is a 'pessimistic' interpretation of the data, and will therefore show a downturn more clearly when the increase of infection rate is slowing.
-
27 May 2020: As a fun little add on, the decoded text version of the genome has been added to the repository.
-
3 Jun 2020: Changed the range for the Growth Ratio plot to provide more detail in the recent data, as it begins to asymptotically approach a value of
1. As such, this graph now only shows data >30 days in. Once the overall event has slowed to 'near completion', the range of this plot will be re-opened to show the growth ratio over the entire course of the event. -
8 Jun 2020: Due to changes in the data presented on the PA Department of Health site, the script is no longer functioning. Apparently they're only updating the ArcGIS site now, and posting that as an iframe in the page, like it's the year 1999 or something. Efforts will be made to obtain the previously available data output from previous days, but until they provide a tabular format, it seems unlikely.
-
9 Jun 2020: Partial restore of the previously available data. Until the DoH restores the county-by-county table, no fully verbose info can be provided. However, info on when the pages have been updated (for manual retrieval) as well as a rough total case count is available. The DoH frequently changes the formatting of this number throughout the day, so it may contain newline and extraneous characters. They never said they were computer experts…
-
20 Jun 2020: The Dept of Health is no longer updating the pages over the weekends. This means that the numbers obtained via the script will not reflect the actual data, and must be manually calculated from the ArcGIS map. Obviously, this is not an ideal situation, but as the numbers wane, it is an expected change. All attempts will be made to accurately show the numbers as updated from the page content. This only further emphasizes the need for repositories of data such as this one.
* Map, table and case count last updated at 12:00 p.m. on 3/26/2020
1,687 cases confirmed statewide
16 deaths confirmed statewide
48 of 67 counties affected
Adams county: 7 cases, 0 deaths
Allegheny county: 133 cases, 2 deaths
Armstrong county: 1 cases, 0 deaths
Beaver county: 13 cases, 0 deaths
Berks county: 36 cases, 0 deaths
Blair county: 1 cases, 0 deaths
Bradford county: 2 cases, 0 deaths
Bucks county: 107 cases, 0 deaths
Butler county: 19 cases, 1 deaths
Cambria county: 1 cases, 0 deaths
Carbon county: 2 cases, 0 deaths
Centre county: 9 cases, 0 deaths
Chester county: 84 cases, 0 deaths
Clearfield county: 2 cases, 0 deaths
Columbia county: 3 cases, 0 deaths
Crawford county: 1 cases, 0 deaths
Cumberland county: 15 cases, 0 deaths
Dauphin county: 13 cases, 0 deaths
Delaware county: 156 cases, 1 deaths
Erie county: 4 cases, 0 deaths
Fayette county: 8 cases, 0 deaths
Franklin county: 5 cases, 0 deaths
Greene county: 3 cases, 0 deaths
Indiana county: 1 cases, 0 deaths
Juniata county: 1 cases, 0 deaths
Lackawanna county: 28 cases, 2 deaths
Warning: 21 active cases in Lancaster county.
Lancaster county: 21 cases, 0 deaths
Lawrence county: 1 cases, 0 deaths
Lebanon county: 4 cases, 0 deaths
Lehigh county: 63 cases, 1 deaths
Luzerne county: 36 cases, 1 deaths
Lycoming county: 1 cases, 0 deaths
Mercer county: 3 cases, 0 deaths
Monroe county: 67 cases, 2 deaths
Montgomery county: 282 cases, 2 deaths
Montour county: 4 cases, 0 deaths
Northampton county: 56 cases, 3 deaths
Philadelphia county: 402 cases, 1 deaths
Pike county: 15 cases, 0 deaths
Potter county: 1 cases, 0 deaths
Warning: 9 active cases in Schuylkill county.
Schuylkill county: 9 cases, 0 deaths
Somerset county: 2 cases, 0 deaths
Susquehanna county: 1 cases, 0 deaths
Warren county: 1 cases, 0 deaths
Washington county: 12 cases, 0 deaths
Wayne county: 6 cases, 0 deaths
Westmoreland county: 24 cases, 0 deaths
York county: 21 cases, 0 deaths
Contributions are welcome. Please ensure that any modifications to the database methods are tested, and validated against a freshly initialized copy of the DB.
PR's related to improving the accuracy and precision of the Growth Ratio are welcomed.