Changwen Xu, Yuyang Wang, Amir Barati Farimani
Carnegie Mellon University
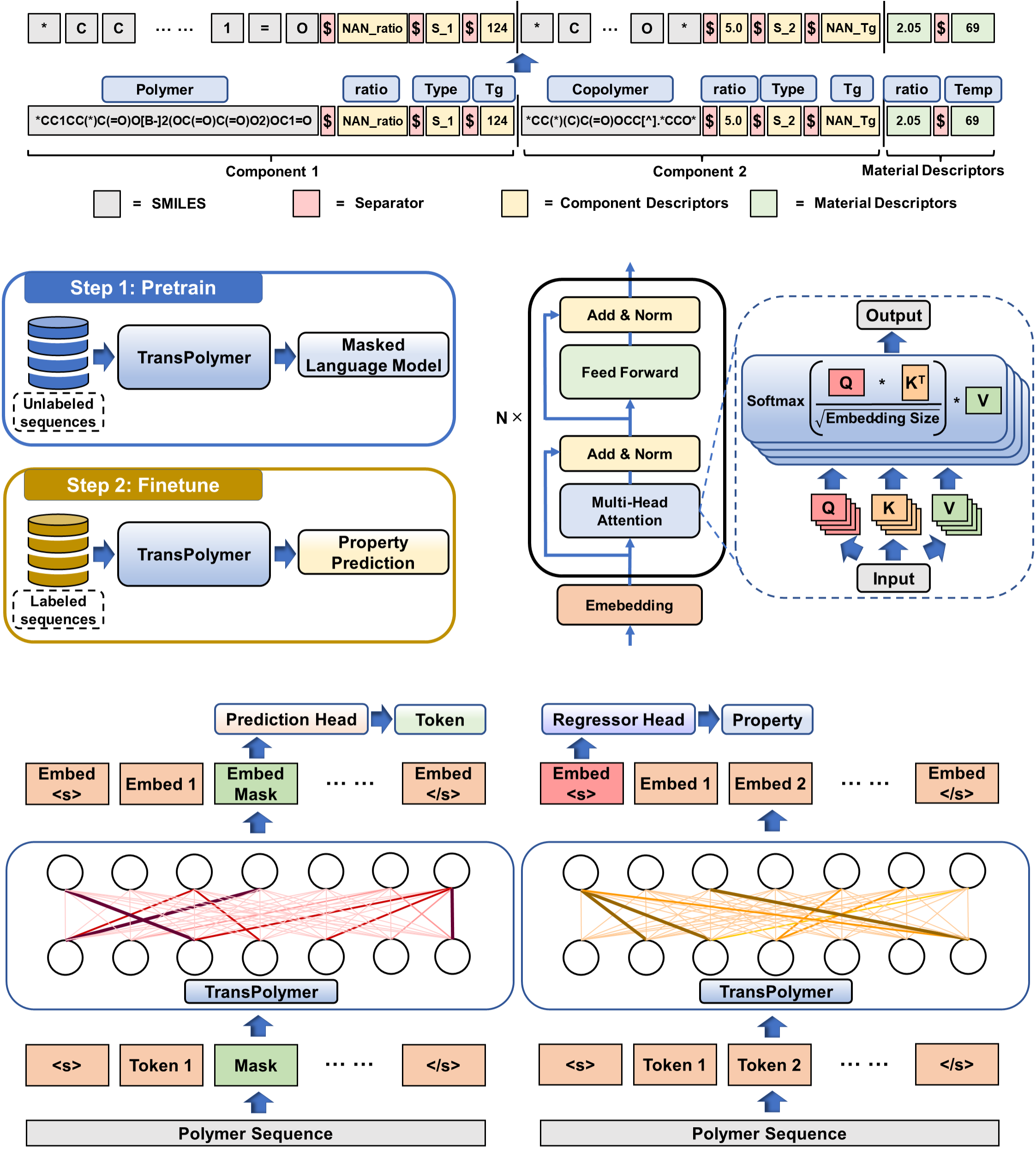
This is the official implementation of TransPolymer: "TransPolymer: a Transformer-based language model for polymer property predictions". In this work, we introduce TransPolymer, a Transformer-based language model, for representation learning of polymer sequences by pretraining on a large unlabeled dataset (~5M polymer sequences) via self-supervised masked language modeling and making accurate and efficient predictions of polymer properties in downstream tasks by finetuning. If you find our work useful in your research, please cite:
@article{xu2023transpolymer,
title={TransPolymer: a Transformer-based language model for polymer property predictions},
author={Xu, Changwen and Wang, Yuyang and Barati Farimani, Amir},
journal={npj Computational Materials},
volume={9},
number={1},
pages={64},
year={2023},
publisher={Nature Publishing Group UK London}
}
Set up conda environment and clone the github repo
# create a new environment
$ conda create --name TransPolymer python=3.9
$ conda activate TransPolymer
# install requirements
$ conda install pytorch==1.12.0 torchvision torchaudio cudatoolkit=11.3 -c pytorch
$ pip install transformers==4.20.1
$ pip install PyYAML==6.0
$ pip install fairscale==0.4.6
$ conda install -c conda-forge rdkit=2022.3.4
$ conda install -c conda-forge scikit-learn==0.24.2
$ conda install -c conda-forge tensorboard==2.9.1
$ conda install -c conda-forge torchmetrics==0.9.2
$ conda install -c conda-forge packaging==21.0
$ conda install -c conda-forge seaborn==0.11.2
$ conda install -c conda-forge opentsne==0.6.2
# clone the source code of TransPolymer
$ git clone https://github.com/ChangwenXu98/TransPolymer.git
$ cd TransPolymer
The pretraining dataset is adopted from the paper "PI1M: A Benchmark Database for Polymer Informatics". Data augmentation is applied by augmenting each sequence to five. Pretraining data with smaller sizes are obtained by randomly picking up data entries from PI1M dataset.
Ten datasets, concerning different polymer properties including polymer electrolyte conductivity, band gap, electron affinity, ionization energy, crystallization tendency, dielectric constant, refractive index, and p-type polymer OPV power conversion efficiency, are used for downstream tasks. Data processing and augmentation are implemented before usage in the finetuning stage. The original datasets and their sources are listed below:
PE-I: "AI-Assisted Exploration of Superionic Glass-Type Li(+) Conductors with Aromatic Structures"
PE-II: "Database Creation, Visualization, and Statistical Learning for Polymer Li+-Electrolyte Design"
Egc, Egb, Eea, Ei, Xc, EPS, Nc: "Polymer informatics with multi-task learning"
The original and processed datasets are included in the data folder.
PolymerSmilesTokenization.py is adapted from RobertaTokenizer from huggingface with a specially designed regular expression for tokenization with chemical awareness.
Pretrained model can be found in ckpt folder.
To pretrain TransPolymer, where the configurations and detailed explaination for each variable can be found in config.yaml.
$ python -m torch.distributed.launch --nproc_per_node=2 Pretrain.py
DistributedDataParallel is used for faster pretraining. The pretrained model can be found in ckpt/pretrain.pt
To finetune the pretrained TransPolymer on different downstream benchmarks about polymer properties, where the configurations and detailed explaination for each variable can be found in config_finetune.yaml.
$ python Downstream.py
To visualize the attention scores for interpretability of pretraining and finetuning phases, where the configurations and detailed explaination for each variable can be found in config_attention.yaml.
$ python Attention_vis.py
To visualize the chemical space covered by each dataset, where the configurations and detailed explaination for each variable can be found in config_tSNE.yaml.
$ python tSNE.py
- PyTorch implementation of Transformer: https://github.com/huggingface/transformers.git