The code for paper " OUCopula: Bi-Channel Multi-Label Copula-Enhanced Adapter-Based CNN for Myopia Screening Based on OU-UWF Images (IJCAI2024 Accepted)’’.
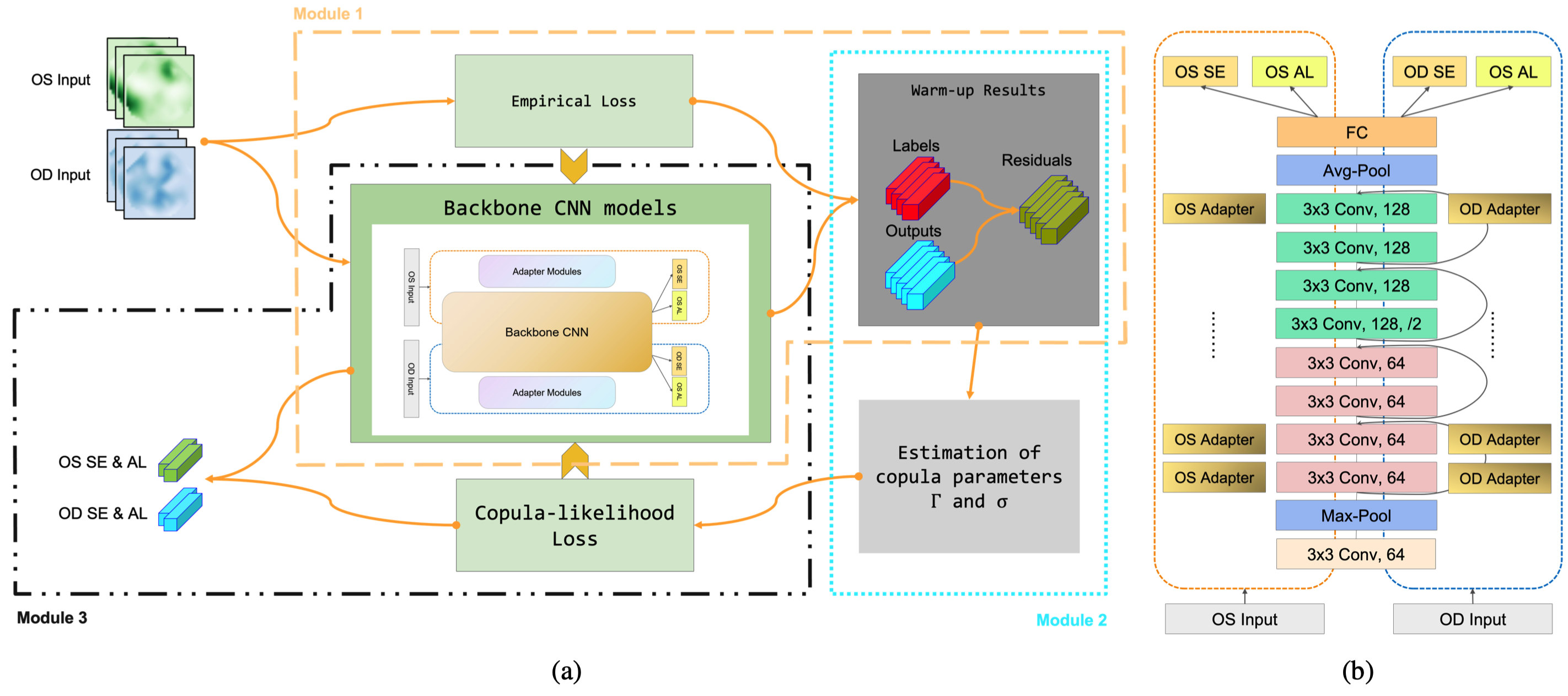
Three modules in OUCopula framework are illustrated in figure (a):
- a warm-up module that trains the backbone under empirical losses,
- a copula estimation module estimates the parameters
$\sigma_{jk}$ and$\gamma_{j_1k_1, j_2k_2}$ in the Gaussian copula based on the results of module 1, - an OUCopula module that trains the backbone CNN using the derived copula-likelihood loss.
The detailed architecture of backbone ResNet with adapter modules is shown in figure (b).
OUCopula requires only a standard computer with enough RAM and a NVIDIA GPU to support operations. We use the following specs:
CPU: 16 vCPU Intel(R) Xeon(R) Platinum 8352V CPU @ 2.10GHz/core
RAM: 90GB
GPU: NVIDIA RTX 4090(24GB)
python == 3.8.10
pandas == 2.0.3
numpy == 1.24.2
aioitertools == 0.11.0
scikit-learn == 1.3.2
torch == 2.0.9+cu118
seaborn == 0.13.0
The UWF fundus images are in JPEG format and compressed to a resolution of 224 x 224 pixels to facilitate subsequent analysis.
The OU UWF fundus images dataset is temporary not available for public access and undergoing a rigorous ethical review process for open sourcing. Once we are authorized, we will make it publicly available.
The training process of OUCopula including three modules:
The training command for module 1 is:
python code/warmup_adapter seed0.ipynbThe best model with lowest validation loss is stored in ./root/autodl-tmp/warmup loader files/WorkID{work_id}_Simu_Warmup_seed{seed}_fold{fold}_X_length{X_length}_batchsize{batch_size}_num_epoch{num_epochs}.pth
and the performance of the best model on test dataset is stored in ./Simu/WorkID{work_id}_Simu_Warmup_seed{seed}_X_length{X_length}_batchsize{batch_size}_num_epoch{num_epochs}.csv.
2. A copula estimation module that estimates the parameters in the Gaussian copula based on the results of the warm-up module
3. An OUCopula module that trains the backbone CNN using the derived copula-likelihood loss that can efficiently capture the dependence structure across multiple labels through a Gaussian copula model
The training command for module 2 and 3 is:
python code/copula_adapter seed0.ipynbThe best model obtained from module 1 is loaded as the pretrained model. The optimal model under copula loss is saved in the /root/autodl-tmp/warmup loader files/WorkID{work_id}_COPULA_seed{seed}_fold{fold}_X_length{X_length}_batchsize{batch_size}_num_epoch{num_epochs}.pth and its performance on the test dataset is saved in the ./Simu/WorkID{work_id}_COPULA_seed{seed}_X_length{X_length}_batchsize{batch_size}_num_epoch{num_epochs}.csv.
The experiment results of OUCopula with backbones "DenseNet" and "ResNet" are reported in summary/Densenet_summary.xlsx and summary/Resnet_summary.xlsx.
This code is for research purposes and is not approved for clinical use.
If you find the codebase useful for your research, please cite our paper:
The citation information will be updated when the official IJCAI24 proceeding is online.
@inproceedings{OUCopula_IJCAI2024,
title = {OUCopula: Bi-Channel Multi-Label Copula-Enhanced Adapter-Based CNN for Myopia Screening Based on OU-UWF Images},
author = {Yang Li, Qiuyi Huang, Chong Zhong, Danjuan Yang, Meiyan Li, A.H. Welsh, Aiyi Liu, Bo Fu, Catherien C. Liu, Xingtao Zhou},
year = {2024},
url = {https://arxiv.org/abs/2403.11974},
doi = {10.48550/arXiv.2403.11974}
}
If you have any questions, please contact us via email: