Follow brew postgresql setup instructions, summarised below
brew install postgresqlrun the following and note the resulting output
ln -sfv /usr/local/opt/postgresql/*.plist ~/Library/LaunchAgents/Users/chrisbailey/Library/LaunchAgents/homebrew.mxcl.postgresql.plist -> /usr/local/opt/postgresql/homebrew.mxcl.postgresql.plist
using the paths output we can run:
alias pg_start="launchctl load /Users/chrisbailey/Library/LaunchAgents/homebrew.mxcl.postgresql.plist"
alias pg_stop="launchctl unload /usr/local/opt/postgresql/homebrew.mxcl.postgresql.plist"verify the installation worked:
createdb `whoami`
psqlpsql (12.2)
\q out of psql and in terminal we run:
\q
createuser -s postgres
createdb testdatabaseand list our databases
psql -U postgres -llist databases:
psql -U postgres -lnavigate to db:
psql testdatabasetables within our db"
\dload in the required packages
library(RPostgreSQL)
library(DBI)
library(dplyr)
library(dbplyr)Init db connection:
con <- RPostgreSQL::dbConnect(RPostgreSQL::PostgreSQL(),
dbname = "testdatabase",
host = 'localhost',
user = "postgres",
password = Sys.getenv('PG_PASS'))set our environment variable with:
Sys.setenv('PG_PASS'='password')
# or
usethis::edit_r_environ()There are two ways we can get a data frame from R into postgreSQL.
- make use of the
DBIfunctiondbWriteTable - write the SQL ourselves and send it to the database with
dbSendQuery
Method 1 looks like this:
data(iris)
dbWriteTable(con, 'iris_1', iris )## [1] TRUE
dplyr::tbl(con, 'iris_1')## # Source: table<iris_1> [?? x 6]
## # Database: postgres 12.2.0 [postgres@localhost:5432/testdatabase]
## row.names Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <chr> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with more rows
Method 2 looks like this (using glue sql):
library(glue)
# create the table schema
DBI::dbSendQuery(con, 'CREATE TABLE IF NOT EXISTS iris_2 (id SERIAL PRIMARY KEY,
sepal_length NUMERIC, sepal_width NUMERIC, petal_length NUMERIC,
petal_width NUMERIC, species VARCHAR(15));')## <PostgreSQLResult>
# set up query with glue sql to insert values line by line
iris$Species <- as.character(iris$Species)
query <- apply(iris, 1, function(x) glue_sql("INSERT INTO iris_2 (sepal_length, sepal_width, petal_length, petal_width, species) VALUES ({x*})", .con = con))
send <- sapply(query, function(x) DBI::dbSendQuery(con, x))
# DBI::dbSendQuery(con, 'drop table iris_2')
tbl(con, 'iris_2')## # Source: table<iris_2> [?? x 6]
## # Database: postgres 12.2.0 [postgres@localhost:5432/testdatabase]
## id sepal_length sepal_width petal_length petal_width species
## <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with more rows
We can drop the table with:
DBI::dbSendQuery(con, 'drop table iris_2;')## <PostgreSQLResult>
In the example above we already saw how to create a primary key with the PRIMARY KEY command. SERIAL tells the id column to auto-increment with a numeric id.
# create the table schema
DBI::dbSendQuery(con, 'CREATE TABLE IF NOT EXISTS iris_2 (id SERIAL PRIMARY KEY,
sepal_length NUMERIC, sepal_width NUMERIC, petal_length NUMERIC,
petal_width NUMERIC, species VARCHAR(15));')## <PostgreSQLResult>
library(dplyr)
iris_3 <- iris %>% janitor::clean_names() %>% tibble::as_tibble()
# add an id column
iris_3 <- iris_3 %>% mutate(row_number = row_number()) %>%
select(row_number, sepal_length, sepal_width, petal_length, petal_width, species)
# create the table schema
DBI::dbSendQuery(con, 'CREATE TABLE IF NOT EXISTS iris_3 (row_number INTEGER CONSTRAINT row_id PRIMARY KEY,
sepal_length NUMERIC, sepal_width NUMERIC, petal_length NUMERIC,
petal_width NUMERIC, species VARCHAR(15));')## <PostgreSQLResult>
query <- apply(iris_3, 1, function(x) glue_sql("INSERT INTO iris_3 (row_number, sepal_length, sepal_width, petal_length, petal_width, species) VALUES ({x*}) ON CONFLICT DO NOTHING ", .con = con))
send <- sapply(query, function(x) DBI::dbSendQuery(con, x))
tbl(con, 'iris_3')## # Source: table<iris_3> [?? x 6]
## # Database: postgres 12.2.0 [postgres@localhost:5432/testdatabase]
## row_number sepal_length sepal_width petal_length petal_width species
## <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with more rows
Where the primary key constrains the row_number column to be unique.
Where the constraint is violated we can tell postgres what to do using
the ON CONFLICT command. DO NOTHING skips inserting the observation;
DO UPDATE updates the existing observation with the incoming
insertion.
We can check the primary key set on the table by running the following commands in terminal:
psql testdatabase
\d iris_3
Table "public.iris_3"
Column | Type | Collation | Nullable | Default
--------------+-----------------------+-----------+----------+---------
row_number | integer | | not null |
sepal_length | numeric | | |
sepal_width | numeric | | |
petal_length | numeric | | |
petal_width | numeric | | |
species | character varying(15) | | |
Indexes:
"constraint_name" PRIMARY KEY, btree (row_number)Set a new primary key constraint with the name ‘iris_pk’:
DBI::dbSendQuery(con, 'ALTER TABLE iris_1 ADD CONSTRAINT iris_pk PRIMARY KEY ("row.names");')## <PostgreSQLResult>
We can remove the primary key constraint by the constraint name we set:
DBI::dbSendQuery(con, "ALTER TABLE iris_1
DROP CONSTRAINT iris_pk;")## <PostgreSQLResult>
While a primary key essentially does this, we might want to set up a constraint to ensure no records are duplicated if, for example, we are using an auto-incrementing primary key.
# set table schema with auto-incrementing primary key
DBI::dbSendQuery(con, 'CREATE TABLE IF NOT EXISTS population (id SERIAL PRIMARY KEY,
date VARCHAR(4), country VARCHAR(64), population NUMERIC);')
# add constraint so we don't accidently add duplicate records
DBI::dbSendQuery(con, "ALTER TABLE population ADD CONSTRAINT nodup UNIQUE (date, country, population);")
# add data
DBI::dbSendQuery(con, "INSERT INTO population (date, country, population) VALUES ('2019', 'UK', 66.65);")
# add duplicate data
DBI::dbSendQuery(con, "INSERT INTO population (date, country, population) VALUES ('2019', 'UK', 66.65);")Error in postgresqlExecStatement(conn, statement, …) : RS-DBI driver: (could not Retrieve the result : ERROR: duplicate key value violates unique constraint “nodup” DETAIL: Key (date, country, population)=(2019, UK, 66.65) already exists. )
Here we get an error if we try and add in a duplicate observation. We should tell postgres what to do if we get a violation of our constraint:
do nothing on violation: If we want to do nothing on any constraint
violation (primary key or unique constraint) then we don’t need to
specify a specific ON CONSTRAINT:
# try to add data
DBI::dbGetQuery(con, "INSERT INTO population (date, country, population) VALUES ('2019', 'UK', 66.65) ON CONFLICT DO NOTHING;")## Error in postgresqlExecStatement(conn, statement, ...) :
## RS-DBI driver: (could not Retrieve the result : ERROR: relation "population" does not exist
## LINE 1: INSERT INTO population (date, country, population) VALUES ('...
## ^
## )
## NULL
update violation with incoming data:
# try to add data
DBI::dbGetQuery(con, "INSERT INTO population (date, country, population) VALUES ('2019', 'UK', 66.65) ON CONFLICT ON CONSTRAINT nodup DO UPDATE SET (date, country, population) = (EXCLUDED.date, EXCLUDED.country, EXCLUDED.population);")## Error in postgresqlExecStatement(conn, statement, ...) :
## RS-DBI driver: (could not Retrieve the result : ERROR: relation "population" does not exist
## LINE 1: INSERT INTO population (date, country, population) VALUES ('...
## ^
## )
## NULL
update violation with some existing data:
# try to add data
DBI::dbGetQuery(con, "INSERT INTO population (date, country, population) VALUES ('2019', 'UK', 66.65) ON CONFLICT ON CONSTRAINT nodup DO UPDATE SET (date, country, population) = (population.date, population.country, EXCLUDED.population);")## Error in postgresqlExecStatement(conn, statement, ...) :
## RS-DBI driver: (could not Retrieve the result : ERROR: relation "population" does not exist
## LINE 1: INSERT INTO population (date, country, population) VALUES ('...
## ^
## )
## NULL
It is obvious from the previous sections that we can send a sql query
with a combination of glue_sql and dbSendQuery. We can also make use
of dplyr syntax using:
tbl(con, 'iris_3')## # Source: table<iris_3> [?? x 6]
## # Database: postgres 12.2.0 [postgres@localhost:5432/testdatabase]
## row_number sepal_length sepal_width petal_length petal_width species
## <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with more rows
tbl doesn’t return the full database but a sample of the result for
speed purposes. If we want to return the full result so we can use the
object in R for further analysis, we use collect:
tbl(con, 'iris_3') %>%
collect()## # A tibble: 150 x 6
## row_number sepal_length sepal_width petal_length petal_width species
## * <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with 140 more rows
We can use typical dplyr syntax to query our data as an alternative to
dbSendQuery. dplyr will convert our dplyr syntax into SQL code!
tbl(con, 'iris_3') %>%
group_by(species) %>%
summarise(avg_length = mean(sepal_length)) %>%
collect()## # A tibble: 3 x 2
## species avg_length
## * <chr> <dbl>
## 1 virginica 6.59
## 2 versicolor 5.94
## 3 setosa 5.01
We can see what code was run in SQL:
tbl(con, 'iris_3') %>%
group_by(species) %>%
summarise(avg_length = mean(sepal_length)) %>%
show_query()## <SQL>
## SELECT "species", AVG("sepal_length") AS "avg_length"
## FROM "iris_3"
## GROUP BY "species"
Run the following commands (or run in psql in terminal) to initialise postgis on our database:
dbGetQuery(con, 'create extension postgis;')
dbGetQuery(con, 'create extension fuzzystrmatch;')
dbGetQuery(con, 'create extension postgis_tiger_geocoder;')
dbGetQuery(con, 'create extension postgis_topology;')Writing a spatial file to a database from R is best done using the sf
packages and sf::write_sf:
library(sf)
uk_auth <- read_sf('https://opendata.arcgis.com/datasets/d54f953d633b45f5a82fdd3c89b4c955_0.geojson')
write_sf(uk_auth, con, 'uk_authorities')To speed up queries on our spatial table we generally want to set a spatial index:
dbSendQuery(con, 'CREATE INDEX auth_gpx ON uk_authorities USING GIST (geography(geometry))')## <PostgreSQLResult>
Because dplyr doesn’t understand the column with class ‘geometry’, we
cannot really query using the tbl method.
We will have to form the query ourselves, for example to get the local
authority containing the point 51.5082, -0.0759 (Tower of London) we
will have to use glue_sql:
lat <- 51.5082
lng <- -0.0759
query <- glue_sql(
"SELECT * FROM uk_authorities
WHERE ST_INTERSECTS(geometry,ST_SetSRID(ST_MakePoint({lng},{lat}),4326))"
,.con=con)
tower_auth <- read_sf(con, query = query)
tower_auth## Simple feature collection with 1 feature and 6 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -0.07940341 ymin: 51.48599 xmax: 0.009089509 ymax: 51.54469
## epsg (SRID): 4326
## proj4string: +proj=longlat +datum=WGS84 +no_defs
## # A tibble: 1 x 7
## objectid cmlad11cd cmlad11nm cmlad11nmw st_areashape st_lengthshape
## <int> <chr> <chr> <chr> <dbl> <dbl>
## 1 321 E41000321 Tower Ha… " " 19771465. 27476.
## # … with 1 more variable: geometry <MULTIPOLYGON [°]>
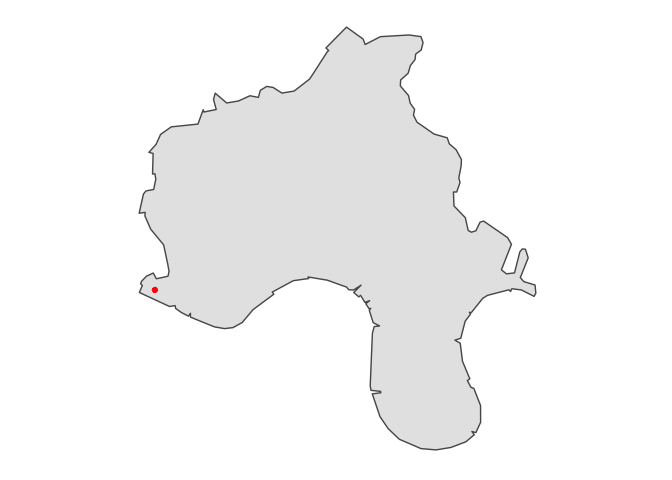
Just for fun, visualise result:
library(ggplot2)
ggplot(tower_auth) +
geom_sf() +
geom_sf(data = st_as_sf(data.frame(lat, lng ), coords = c('lng','lat'), crs=4326), color='red') +
ggthemes::theme_map() cd to the directory we want to save the db dump in and run:
pg_dump --port=5432 --username=postgres --dbname=testdatabase --file=postgresdump.sqlClose the connection:
DBI::dbDisconnect(con)Sometimes we can accumulate multiple connections, often resulting in the error that we’ve opened the maximum number of connections. We can loop through all of them and close them all:
lapply(dbListConnections(PostgreSQL()), dbDisconnect)## [[1]]
## [1] TRUE
Of course it’s best practice to not accumulate connections in the first
place. If using a function that opens a connection each time it’s run,
it’s best to ensure it gets closed using on.exit:
db_fn <- function() {
on.exit(dbDisconnect(con))
con <- RPostgreSQL::dbConnect(RPostgreSQL::PostgreSQL(),
dbname = "testdatabase",
host = 'localhost',
user = "postgres",
password = Sys.getenv('PG_PASS'))
tbl(con, 'iris_3') %>%
collect()
}
db_fn()## # A tibble: 150 x 6
## row_number sepal_length sepal_width petal_length petal_width species
## * <int> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 1 5.1 3.5 1.4 0.2 setosa
## 2 2 4.9 3 1.4 0.2 setosa
## 3 3 4.7 3.2 1.3 0.2 setosa
## 4 4 4.6 3.1 1.5 0.2 setosa
## 5 5 5 3.6 1.4 0.2 setosa
## 6 6 5.4 3.9 1.7 0.4 setosa
## 7 7 4.6 3.4 1.4 0.3 setosa
## 8 8 5 3.4 1.5 0.2 setosa
## 9 9 4.4 2.9 1.4 0.2 setosa
## 10 10 4.9 3.1 1.5 0.1 setosa
## # … with 140 more rows