DeBaCl is a Python library for density-based clustering with level set trees.
Level set trees are a statistically-principled way to represent the topology of a probability density function. This representation is particularly useful for several core tasks in statistics:
- clustering, especially for data with multi-scale clustering behavior
- describing data topology
- exploratory data analysis
- data visualization
- anomaly detection
DeBaCl is a Python implementation of the Level Set Tree method, with an emphasis on computational speed, algorithmic simplicity, and extensibility.
DeBaCl is available under the 3-clause BSD license.
DeBaCl is currently compatible with Python 2.7 only. Other versions may work, but caveat emptor; at this time DeBaCl is only officially tested on Python 2.7. The package can be downloaded and installed from the Python package installer. From a terminal:
$ pip install debaclIt can also be installed by cloning this GitHub repo. This requires updating the Python path to include the cloned repo. On linux, this looks something like:
$ git clone https://github.com/CoAxLab/DeBaCl/
$ export PYTHONPATH='DeBaCl'All of the dependencies are Python packages that can be installed with either conda or pip. DeBaCl 1.0 no longer depends on igraph, which required tricky manual installation.
Langauges:
- Python 2.7
- (coming soon: Python 3.4)
Required packages:
- numpy
- networkx
- prettytable
Strongly recommended packages
- matplotlib
- scipy
Optional packages
- scikit-learn
X = make_moons(n_samples=100, noise=0.1, random_state=19)[0]
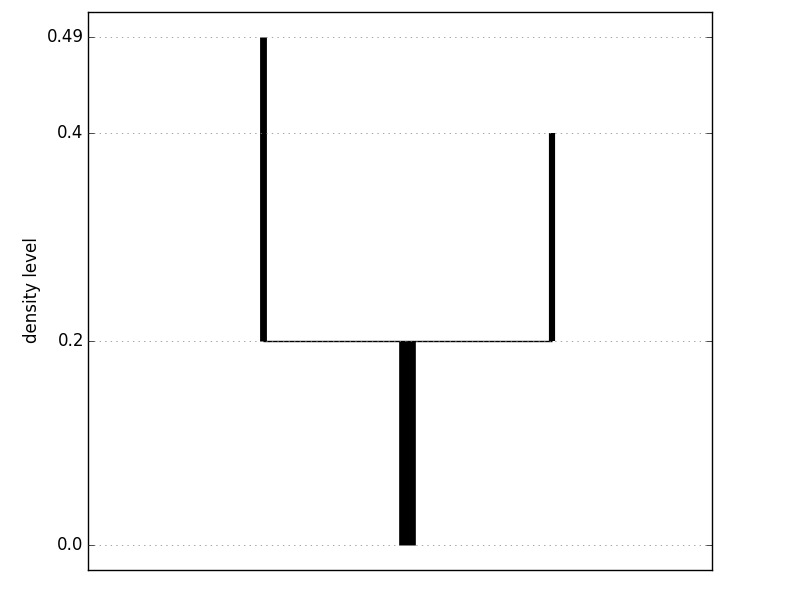
tree = dcl.construct_tree(X, k=10, prune_threshold=10) print tree
```no-highlight
+----+-------------+-----------+------------+----------+------+--------+----------+
| id | start_level | end_level | start_mass | end_mass | size | parent | children |
+----+-------------+-----------+------------+----------+------+--------+----------+
| 0 | 0.000 | 0.196 | 0.000 | 0.220 | 100 | None | [1, 2] |
| 1 | 0.196 | 0.396 | 0.220 | 0.940 | 37 | 0 | [] |
| 2 | 0.196 | 0.488 | 0.220 | 1.000 | 41 | 0 | [] |
+----+-------------+-----------+------------+----------+------+--------+----------+
labels = tree.get_clusters(method='leaf') # each leaf node is a cluster clusters = X[labels[:, 0], :]
fig, ax = plt.subplots() ax.scatter(X[:, 0], X[:, 1], c='black', s=40, alpha=0.4) ax.scatter(clusters[:, 0], clusters[:, 1], c=labels[:, 1], s=80, alpha=0.9, cmap=plt.cm.winter) ax.set_xlabel('x0') ax.set_ylabel('x1', rotation=0) fig.show()
<!---->
<img src="docs/readme_clusters.png" height="480px" />
Documentation
-------------
- [API documentation](http://debacl.readthedocs.io/en/master/)
- [Example jupyter notebooks](examples) (in progress)
Running unit tests
------------------
From the top level of the repo:
```bash
$ nosetests -s -v debacl/test
-
Chaudhuri, K., & Dasgupta, S. (2010). Rates of Convergence for the Cluster
Tree. In Advances in Neural Information Processing Systems 23 (pp. 343–351). Vancouver, BC. -
Kent, B. P., Rinaldo, A., Yeh, F.-C., & Verstynen, T. (2014). Mapping Topographic Structure in White Matter Pathways with Level Set Trees. PLoS ONE.
-
Kent, B. P., Rinaldo, A., & Verstynen, T. (2013). DeBaCl: A Python Package for Interactive DEnsity-BAsed CLustering. arXiv preprint:1307.8136.
-
Kent, B.P. (2013). Level Set Trees for Applied Statistics.