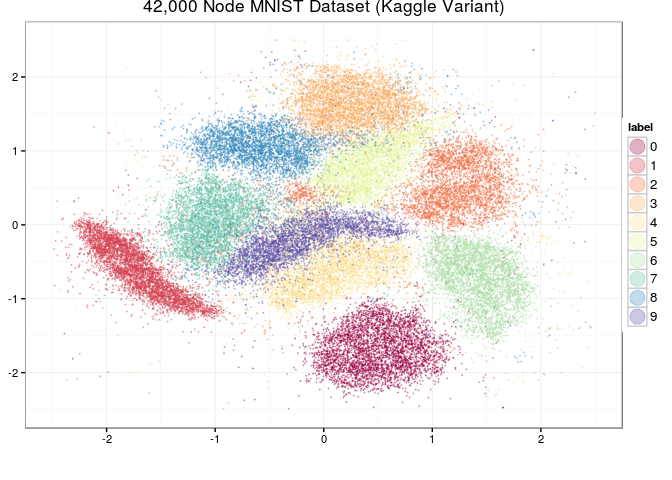
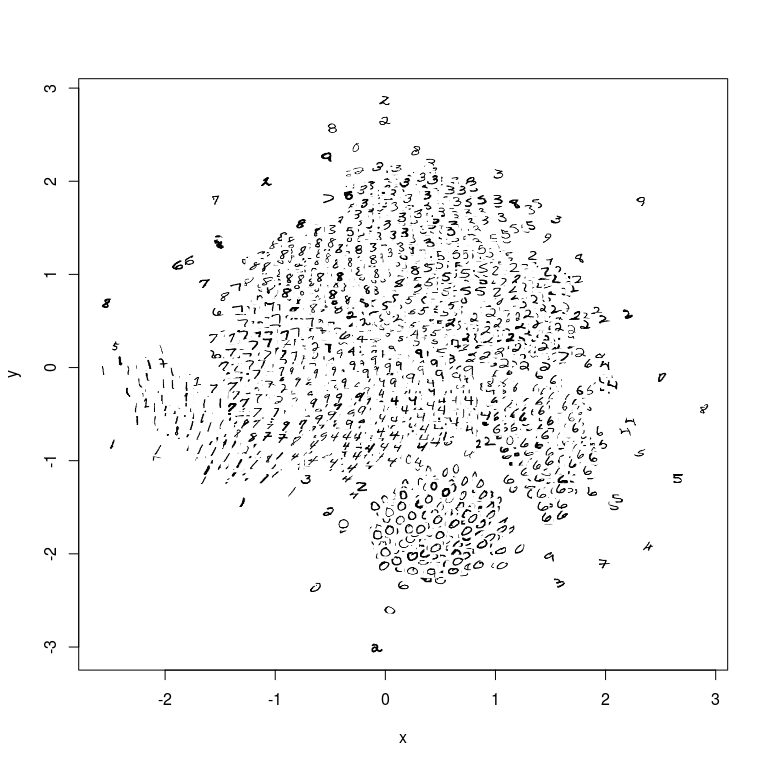
This is an implementation of the largeVis algorithm described in (https://arxiv.org/abs/1602.00370). It also incorporates code for a very fast algorithm for estimating k-nearest neighbors, and for visualizing a map of the manifold.
The inner loops for nearest-neighbor search and gradient descent are implemented in C++ using Rcpp and RcppArmadillo.
- It works!
- The version originally uploaded had issues with OpenMP on some systems. While I work on fixing that, OpenMP is disabled in the current version. Other changes:
- The input matrix to the
randomProjectionTreeSearchfunction should now be transposed so examples are columns and rows are features. - The alternative distance function where α = 0 is partially implemented.
- Progress now depends on
RcppProgressinstead ofprogress. These progress reports are substantially less pretty -- but much faster, and theprogresspackage was causing crashes in some cases.
- The input matrix to the
- The map visualization works, but is only tested for the case where the images are presented as an array of greyscale images.
- This project is under heavy development.
- I am attempting to replicate the paper's results with larger and larger datasets. This takes time because my hardware is not as powerful as the authors'. If you have any to volunteer, please contact me!
- The algorithm is memory intensive. Processing mnist, memory usage peaked at approximately 8GB. I would appreciate any reports using it with larger datasets.
- Note that your installation of R must be configured to work with OpenMP. I have had a report that on Federa 22, even small datasets could not be processed because of exceeding the C stack space. If you experience any compilation issues or similar crashes, please create an issue.
load("./mnist.Rda")
dat <- mnist$images
dim(dat) <- c(42000, 28 * 28)
dat <- (dat / 255) - 0.5
dat <- t(dat)
coords <- vis(dat, check=FALSE,
n_tree = 50, tree_th = 100,
K = 50, alpha = 2, max.iter = 4)flip <- function(x) apply(x,2,rev)
rotate <- function(x) t(flip(x))
mnistimages <- apply(mnist$images,
MARGIN=1,
FUN = function(x) as.array(rotate(flip(x))))
mnistimages <- t(mnistimages)
dim(mnistimages) <- c(42000, 28, 28)
coords <- as.matrix(coords[,1:2])
coords <- scale(coords)
manifoldMap(coords,
n = 3000,
scale = 0.005,
transparency = F,
images = mnistimages,
xlab=NULL, ylab=NULL)