Cosmology sampling with Population Monte Carlo (PMC)
CosmoPMC is a Monte-Carlo sampling method to explore the likelihood of various cosmological probes. The sampling engine is implemented with the package pmclib. It is called Population MonteCarlo (PMC), which is a novel technique to sample from the posterior Capp'{e} et al. 2008. PMC is an adaptive importance sampling method which iteratively improves the proposal to approximate the posterior. This code has been introduced, tested and applied to various cosmology data sets in Wraith, Kilbinger, Benabed et al.(2009). Results on the Bayesian evidence using PMC are discussed in Kilbinger, Wraith, Benabed et al. (2010).
Martin Kilbinger
Karim Benabed, Olivier Cappé, Jean Coupon, Jean-François Cardoso, Gersende Fort, Henry Joy McCracken, Simon Prunet, Christian P. Robert, Darren Wraith
1.4
CosmoPMC requires the libraries
nicaea,
pmclib, and third-party libraries and
programs such as gsl, fftw3, lacpack, or cmake. Download and run the
automatic script install_CosmoPMC.sh to build all
dependent packages and programs into a conda virtual environment. The only
prerequisite (apart from the bash shell) is conda, which can be downloaded
and installed from
https://docs.conda.io/en/latest/miniconda.html.
Once conda is installed and in the search path, the installation of CosmoPMC should be easy:
git clone https://github.com/CosmoStat/CosmoPMC
cd CosmoPMC
./install_CosmoPMC.sh --no-R [OPTIONS]Type ./install_CosmoPMC -h for help.
You might need to activate the cosmopmc conda environment after installation, with
conda activate cosmopmcOn success, the command line prompt should start now with the string (cosmopmc).
You can also install all packages by hand. First, download and install the CosmoPMC-adjacent packages, from their respective github pages for nicaea and pmclib.
Next, if not alreay done, download the CosmoPMC package from the github repository:
git clone https://github.com/CosmoStat/CosmoPMCA new directory CosmoPMC will be created automatically. Change into that directory, and configure the code with the (poor-man's) python configuration script.
cd CosmoPMC
./configure.py [FLAGS]You will need to indicate paths to libraries and other flags. At the minimum, you probably need to specify the basic paths to the libraries nicaea and pmclib. (Specify both even if the paths are the same). Type ./configure.py -h to see all options.
After configuration, compile the code as follows:
makeIf you need to topolike external module, the following steps are required.
-
Compile the
topolikecode and create the `topotest' test program. -
Create the topolike library. In the
topolikecode directory:
ar rv libtopo.a *.o
- On some computing architectures, the linker flags need to be communicated to
CosmoPMC. This can be done by using the option--lflags LFLAGSforinstall_CosmoPMC.sh, and setting all flags asLFLAGS.
See the directory Demo/tempering and the corresponding readme.
To get familiar with CosmoPMC, use the examples which are included
in the software package. Simply change to one of the subdirectories in
Demo/MC_Demo and proceed on to the subsection
Run below. A quick-to-run likelihood is the supernova one, in Demo/MC_Demo/SN.
To run different likelihood combinations, using existing or your own data, the following two steps are recommended to set up a CosmoPMC run.
- Data and parameter files
Create a new directory and copy data files. You can do this automatically for the pre-defined
probes of CosmoPMC by using
newdir_pmc.shWhen asked, enter the likelihood/data type. More than one type can be chosen by
adding the corresponding (bit-coded) type id’s. Symbolic links to corresponding
files in COSMOPMC/data are set, and parameter files from COSMOPMC/par_files
are copied to the new directory on request.
- Configuration file
Create the PMC configuration file config_pmc. Examples for existing data modules
can be found in COSMOPMC/Demo/MC_Demo. In some cases, information about
the galaxy redshift distribution(s) have to be provided, and the corresponding
files (nofz*) copied. See Examples above.
Type
/path/to/CosmoPMC/bin/cosmo pmc.pl -n NCPUto run CosmoPMC on NCPU CPUs. See cosmo pmc.pl -h for more options.
Depending on the type of initial proposal, a maximum-search is started followed by a
Fisher matrix calculation. After that, PMC is started.
Depending on the machine's architecture, the default way to use MPI (calling the executable with mpirun) might not be supported. In that case you will have to run the PMC part by using the executable /path/to/CosmoPMC/bin/cosmo pmc, or modifying the perl script.
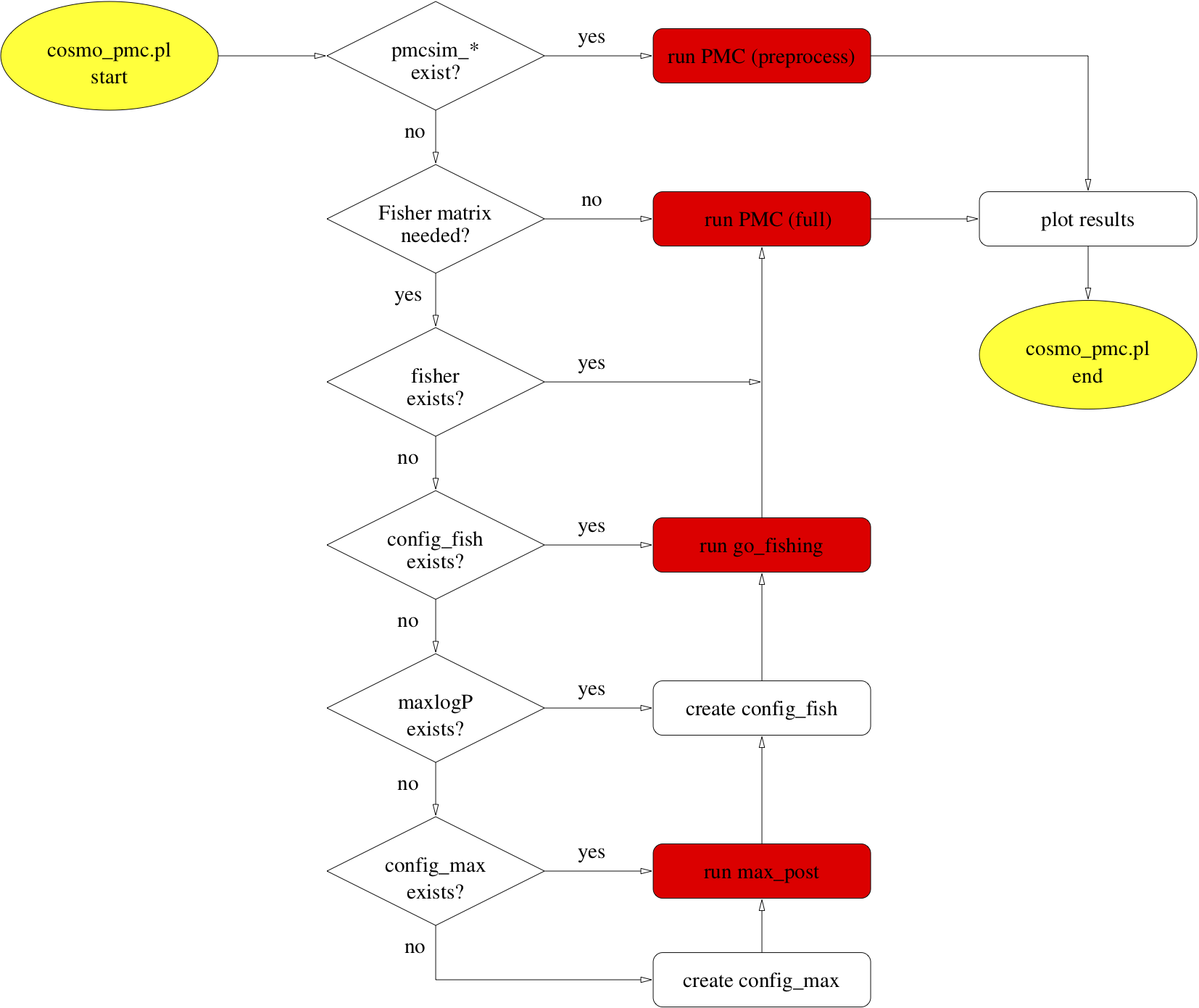
The figure below shows a flow chart of the script’s actions.
Check the text files perplexity and enc. If the perplexity reaches values of 0.8 or
larger, and if the effective number of components (ENC) is not smaller than around
1.5, the posterior has very likely been explored sufficiently. Those and other
files are being updated during run-time and can be monitored while PMC is running.
The results are stored in the subdirectory of the last, final PMC iteration,
iter_{niter-1}/. The text file mean contains mean and confidence levels.
The file all_cont2d.pdf (when R is used, or all_contour2d.pdf for yorick+perl)
shows plots of the 1d- and 2d-marginals. Plots can be
redone or refined, or created from other than the last iteration with
plot_confidence.R (or plot_contour2d.pl), both scripts are in /path/to/CosmoPMC/bin.
To have cosmo_pmc.pl create these plots, the program R (or yorick) have to be installed.
For R, also install the libraries coda, getopt, and optparse.
Note that in the default setting the posterior plots are not smoothed, this can be achieved
using various command line options, see plot_confidence.R -h (or plot_contour2d.pl -h).
Check out the latest version of the manual
The manual for v1.2 can be found on arXiv, at http://arxiv.org/abs/1101.0950.
CosmoPMC is also listed in ASCL at ascl:1212.006.
If you use CosmoPMC in a publication, please cite the last paper in the list below (Wraith, Kilbinger, Benabed et al. 2009).
Kilbinger et al. (2011): Cosmo Population Monte Carlo - User's manual. Note that earlier version of CosmoPMC <=1.2) contain pmclib and nicaea as built-in code instead of external libraries.
Kilbinger, Benabed et al. (2012): ASCL link of the software package
Kilbinger, Wraith, Benabed et al. (2010): Bayesian evidence
Wraith, Kilbinger, Benabed et al. (2009): Comparison of PMC and MCMC, parameter estimation. The first paper to use CosmoPMC.