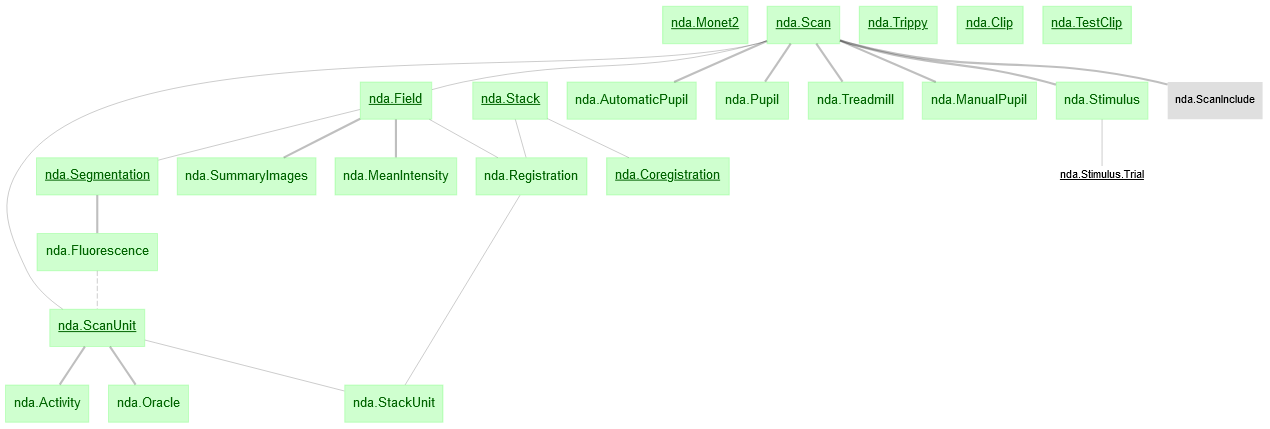
nda schema for MICrONS phase3
This package requires PyTorch. Windows users may need to install from the PyTorch website: PyTorch installation
This package requires the em_coregistration package from the Allen Institute:
pip3 install git+https://github.com/AllenInstitute/em_coregistration.git@phase3Install this package:
pip3 install git+https://github.com/cajal/microns_phase3_nda.gitImport datajoint. Configuration instructions: https://docs.datajoint.io/python/setup/01-Install-and-Connect.html
import datajoint as djIn a jupyter notebook:
from phase3 import nda, func, utilsTo view schema ERD:
dj.ERD(nda)For tutorial see: notebook example
nda.Scan: Information on completed scans. Cajal Pipeline: meso.ScanInfo
nda.Pupil: Pupil traces for each scan. Cajal Pipeline: pupil.py
nda.Treadmill: Treadmill for each scan. Cajal Pipeline: treadmill.py
nda.Stimulus: For each scan, contains the movie aligned to activity traces in nda.Activity.
nda.Stimulus.Trial: Part table of nda.Stimulus. Contains information for each trial. There are three types of trials, Clip, Monet2, and Trippy. Each unique trial has its own condition_hash. To get detailed information for each trial stimulus, join each condition_hash according to its corresponding type in one of: nda.Clip, nda.Monet2, or nda.Trippy.
nda.Clip: Detailed information for movie clips.
nda.Monet2: Detailed information for the Monet2 stimulus.
nda.Trippy Detailed information for the Trippy stimulus.
nda.Field: Individual fields of scans. Cajal Pipeline: meso.ScanInfo.Field
nda.SummaryImages Reference images of the scan field. Cajal Pipeline: meso.SummaryImages
nda.Stack: High-res anatomical stack information. Cajal Pipeline: stack.CorrectedStack
nda.Registration: Parameters of the affine matrix learned for field registration into the stack. Cajal Pipeline: stack.Registration.Affine
nda.Coregistration: Coregistration transform solutions from the Allen Institute. em_coregistration
nda.Segmentation: CNMF segmentation of a field. It records the masks of all segmented cells. Mask_id's are unique per field. Cajal Pipeline: meso.Segmentation.Mask
nda.Fluorescence: Records the raw fluorescence traces for each segmented mask. Cajal Pipeline: meso.Fluorescence.Trace
nda.ScanUnit: Unit_id assignment that is unique across the entire scan. Includes info about each unit. Cajal Pipeline: meso.ScanSet.Unit / meso.ScanSet.UnitInfo
nda.Activity: Deconvolved spike trace from the fluorescence trace. Cajal Pipeline: meso.Activity.Trace
nda.Oracle: Leave-one-out correlation for repeated videos in stimulus.
nda.StackUnit: Unit coordinates in stack reference frame after field registration. np_x, np_y, np_z should be used for transformation to EM space using Coregistration. meso.StackCoordinates.UnitInfo
For more documentation see: Cajal Pipeline Documentation
nda.ScanInclude Scans determined suitable for analysis.
nda.MeanIntensity # Mean intensity of imaging field over time. Cajal Pipeline: meso.Quality.MeanIntensity
Supported by the Intelligence Advanced Research Projects Activity (IARPA) via Department of Interior / Interior Business Center (DoI/IBC) contract number D16PC00003. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. Disclaimer: The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of IARPA, DoI/IBC, or the U.S. Government.