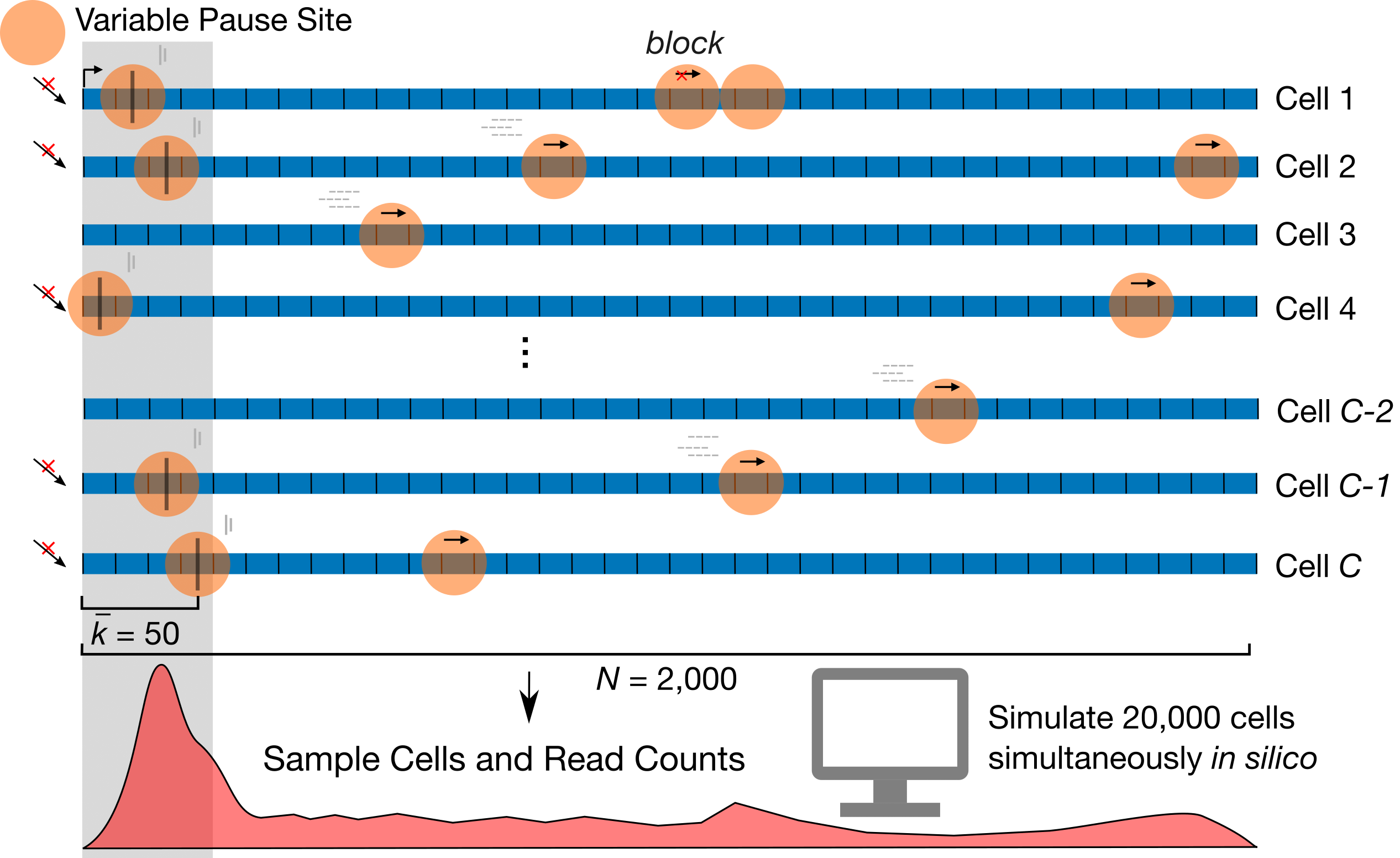
SimPol tracks the independent movement of RNAPs along the DNA templates of a large
number of cells. It accepts several key
user-specified parameters, including the initiation rate, pause-escape rate,
a constant or variable elongation rate, the mean and variance of pause sites across cells,
as well as the center-to-center spacing constraint between RNAPs,
the number of cells being simulated, the gene length, and the total time of transcription.
The simulator simply allows each RNAP to move forward or not,
in time slices of
Fig.1 Design of SimPol (“Simulator of Polymerases”)
We provide three different approaches to set up the environment for SimPol, using either Singularity or Conda or Docker. Pre-built debug and release executables can be found in the bin folder
wget http://compgen.cshl.edu/simpol/simpol.sif
singularity shell simpol.sif
source activate simpol
./bin/simPol_Release --help
Note: Use command below to build container from Singularity file
singularity build --fakeroot simpol.sif Singularity
docker pull cshlhassett/simpol
sudo docker run -it cshlhassett/simpol
source activate simpol
./SimPolv2/bin/simPol_Release --help
conda env create -f environment.yml
conda activate simpol
# Follow commands below to build from source and run
There is the option to build in either debug mode with debug symbols or release mode with compiler optimizations (e.g. -O2).
To create a build directory in release mode
cmake -S . -B Release/ -D CMAKE_BUILD_TYPE=Release
To clean the release mode build directory
cmake --build Release/ --target clean
To build in release mode
cmake --build Release/
Note: Replace all instances of 'Release' with 'Debug' to build in Debug mode
Set Number of Threads [By default uses all available threads]
export OMP_NUM_THREADS=<number of threads to use>
Run in Release Mode
./Release/simPol [options]
Run in Debug Mode
./Debug/simPol [options]
Run program with file containing command line arguments
./Release/simPol $(<simPol_arguments.dat)
qsub -pe OpenMP 5 -cwd -b y ./Release/simPol -n 100 --csv 10
- Note: This allocates 5 threads for the program in the OpenMP parallel environment
- Check available parallel environments: qconf -spl
Options:
-h, --help
Show this help message and exit
-k INTEGER, --tssLen=INTEGER
define the mean of pause sites across cells [default 50bp]
--kSd=DOUBLE
define the standard deviation of pause sites across cells [default 0]
--kMin=INTEGER
upper bound of pause site allowed [default 17bp]
--kMax=INTEGER
lower bound of pause site allowed [default 200bp]
--geneLen=INTEGER
define the length of the whole gene [default 2000bp]
-a DOUBLE, --alpha=DOUBLE
initiation rate [default 1 event per min]
-b DOUBLE, --beta=DOUBLE
pause release rate [default 1 event per min]
-z DOUBLE, --zeta=DOUBLE
the mean of elongation rates across sites [default 2000bp per min]
--zetaSd=DOUBLE
the standard deviation of elongation rates across sites [default 1000]
--zetaMax=DOUBLE
the maximum of elongation rates allowed [default 2500bp per min]
--zetaMin=DOUBLE
the minimum of elongation rates allowed [default 1500bp per min]
--zetaVec=CHARACTER
a file contains vector to scale elongation rates. All cells share the same set of parameters [default ""]
-n INTEGER, --cellNum=INTEGER
Number of cells being simulated [default 10]
-s INTEGER, --polSize=INTEGER
Polymerase II size [default 33bp]
--addSpace=INTEGER
Additional space in addition to RNAP size [default 17bp]
-t DOUBLE, --time=DOUBLE
Total time of simulating data in a cell [default 0.1 min]
--hdf5=INTEGER
Record position matrix to HDF5 file for remaining number of steps specified [default: 0 steps]
--csv=INTEGER
Record position matrix to csv file for remaining number of steps specified [default: 1 step]
-d CHARACTER, --outputDir=CHARACTER
Directory for saving results [default: 'results']
After simulation, SimPol produces multiple output files:
- probability_vector.csv
- pause_sites.csv
- combined_cell_data.csv: Stores the total # of polymerase at each site across all cells
- position_matrices.h5: An HDF5 file containing a group of position matrices for the last number of specified steps. The file can be viewed by importing it into the HDFView app. Download Here: https://www.hdfgroup.org/downloads/hdfview/#download
- Using HighFive interface to generate HDF5 file https://github.com/BlueBrain/HighFive
- Uses chunking and ZLIB (deflate) compression at level 9
- positions/position_matrix_#.csv: CSV files containing the position matrix at a specified step
If you prefer to simulate nascent RNA sequencing read counts in addition to the RNAP positions,
you can also follow the tutorial in scripts/sample_read_counts.Rmd.
Zhao, Y., Liu, L. & Siepel, A. Model-based characterization of the equilibrium dynamics of transcription initiation and promoter-proximal pausing in human cells. 2022.10.19.512929 Preprint at bioRxiv (2022).