Medical data is extremely hard to find due to HIPAA (Health Insurance Portability and Accountability Act) privacy regulations. As a group, we decided to dip our feet into the fast growing field of Machine Learning and NLP (Natural Language Processing) to extract valuable information from unstructed medical text data. Why NLP in healthcare? A large part of medical information is reported as free-text and patient clinical history has been conveyed for centuries by all medical professionals in the form of notes, reports, trascriptions.
There are many applications of text analysis in healthcare. One of the many is Healthcare Records Analysis: combining patient records with their biometric data allows hospitals to identify high-risk patients based on their ongoing treatment records. Many companies like Evidation, Tempus, etc. and huge research centers (Icahn Medical school) specialize in taking healthcare records in a variety of ways to uncover advanced health insights. This field is called “precision medicine”
For our final project, our group chose to use a dataset (from Kaggle) that contained medical transcriptions and the respective medical specialties (4998 datapoints). We chose to implement multiple supervised classification machine learning models - after heavily working on the corpora - to see if we were able to correctly classify the medical specialty based on the trascription text.
- CSV File
- Pandas
- Numpy
- Matplotlib
- Scikit-learn
- NLTK (Natural Language Toolkit)
- RandomForest
- Multinomial Naive Bayes
- LogisticRegression
- Hyperparameter tuning with GridSearchCV (RandomForest)
- TfidfVectorizer
- Yellowbrick
- Plotly
- Gensim & Doc2Vec
- PCA
- Gensim
Preprocessing Data
Our project has three notebooks which shows the different phases of our work as far as cleaning, ideas, and analysis.
Our method was a constant give and take - trying different methods, seeing how they perfomed, and adjusting based on what we saw.
Across all notebooks cleaning the data set and getting rid of characters in the raw text even before the tokenization.

Notebook 1 Classic Approach
- Character lowercase and removal: removed specific characters in our medical transcription column
- Word Tokenization: split our transcription sentences into smaller parts (individual words) called tokens
- Stopwords: removed 'stopwords' (words that provide no meaning to sentence)
- Dropped / Combined Medical Specialties: removed specialties not needed (workers comp, hospice, etc.) / combined specialties together
- Lemmatized words : converted our tokenized words into its meaningful base form
- Limit the lenght of our corpora
- POS Tagging: marked up words with specific part of speech
Notebook 2 Medical Approach From Notebook 2, we gave more importance to Nouns on the assumptions that the Medical vocabolary Nouns are specific to this field and they carry the true meaning. We also were more agressive and removed dates, numbers, PHI etc.
- We eliminated more characters
- Performed POS Tagging before lemmatizing the words to remove all POS tags that were not Nouns
-
- Didn't take away English Stop Words (ex. English words like 'back' are treated as stop words, but in medical those field might be useful)
Notebook 3 Random under-sampling
Machine Learning algorithms tend to produce unsatisfactory classifiers when faced with imbalanced datasets. Standard classifier algorithms like Decision Tree and Logistic Regression have a bias towards classes which have number of instances. They tend to only predict the majority class data. The features of the minority class are treated as noise and are often ignored. Thus, there is a high probability of misclassification of the minority class as compared to the majority class. Dealing with imbalanced datasets like ours entails strategies such as improving classification algorithms or balancing classes in the training data (data preprocessing) before providing the data as input to the machine learning algorithm. The later technique is preferred as it has wider application. The main objective of balancing classes is to either increasing the frequency of the minority class or decreasing the frequency of the majority class. This is done in order to obtain approximately the same number of instances for both the classes. There are a few resampling techniques. In our case, we felt that Random Under-Sampling was appropriare. Random Undersampling aims to balance class distribution by randomly eliminating majority class examples. This is done until the majority and minority class instances are balanced out. We reduced the amount of datapoints of the majority classes (Surgery and Consultation) with the sample() function and combine some of the categories with lower amount od datapoints into the specialty 'Others'. The Disanvantage of this approach is that we discarded potentially useful information which could have be important for building rule classifiers and samples chosen by random under sampling may have been a biased sample and not an accurate representative of the population. Thereby, resulting in inaccurate results with the actual test data set. However, running our models multiple times the scores obtained were always close to each other and the model saved always above the best score of 40%.
Machine learning algorithms cannot work with raw text directly; the text must be converted into numbers. Specifically, vectors of numbers. This is called feature extraction or feature encoding.
- A popular and simple method of feature extraction with text data is called the bag-of-words model of text. A bag-of-words is a representation of text that describes the occurrence of words within a document. It is called a “bag” of words, because any information about the order or structure of words in the document is discarded. The model is only concerned with whether known words occur in the document and how to score the presence of known words, not where these words are in the document. There are four types of vector encoding—frequency, one-hot, TF–IDF, and distributed representations and they can be implemeneted in Scikit-Learn, Gensim, and NLTK. The choice of a specific vectorization technique will be largely driven by the problem space. In this project we used TfidfVectorizer, that has as central insights that meaning is most likely encoded in the more rare terms from a document. Under the hood, the TfidfVectorizer uses the CountVectorizer estimator followed by a TfidfTransformer, which normalizes the occurrence counts by the inverse document frequency.
- Algorithms used: NaiveBayes (Multinomial), RandomForest, Hyperparameter tuning with GridSearchCV (RandomForest), Logistic Regression.
Limitations: One benefit of TF–IDF is that it naturally addresses the problem of stopwords, those words most likely to appear in all documents in the corpus (e.g., “a,” “the,” “of”, etc.), and thus will accrue very small weights under this encoding scheme. This biases the TF–IDF model toward moderately rare words. The bag_of_words model ignores the context, and in turn meaning of words in the document (semantics). Context and meaning can offer a lot to the model, that if modeled could tell the difference between the same words differently arranged (“this is interesting” vs “is this interesting”), synonyms (“old bike” vs “used bike”), and much more.
 Tokens #: after some reduction in specialties (8) and before deciding to put a lowerbound and upperbound for their #
Tokens #: after some reduction in specialties (8) and before deciding to put a lowerbound and upperbound for their #
 Lemmatized # (With Reduction, before deciding to reduce Surgery and Consultation )
Lemmatized # (With Reduction, before deciding to reduce Surgery and Consultation )
Precision can be seen as a measure of a classifier’s exactness. Said another way, “for all instances classified positive, what percent was correct?”
Recall is a measure of the classifier’s completeness; the ability of a classifier to correctly find all positive instances. Said another way, “for all instances that were actually positive, what percent was classified correctly?”
F1 score is a weighted harmonic mean of precision and recall such that the best score is 1.0 and the worst is 0.0. Generally speaking, F1 scores are lower than accuracy measures as they embed precision and recall into their computation. As a rule of thumb, the weighted average of F1 should be used to compare classifier models, not global accuracy.
Support is the number of actual occurrences of the class in the specified dataset. Imbalanced support in the training data may indicate structural weaknesses in the reported scores of the classifier and could indicate the need for stratified sampling or rebalancing.
As it can be easily seen on the classification report heatmaps, Hyperparameter tuning with GridSearchCV on the RandomForest gave us the best result. Note that for Dentistry, we had very good results before blending it to the category "Others", most probably due to the very sectorial vocabulary of a dentist.
 Confusion Matrix (without reduction)
Confusion Matrix (without reduction)
 Confusion Matrix (with reduction)
Confusion Matrix (with reduction)
** Doc2Vec with Gensim **
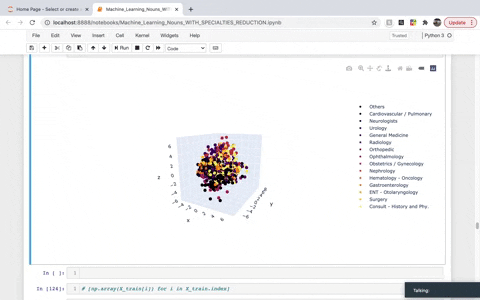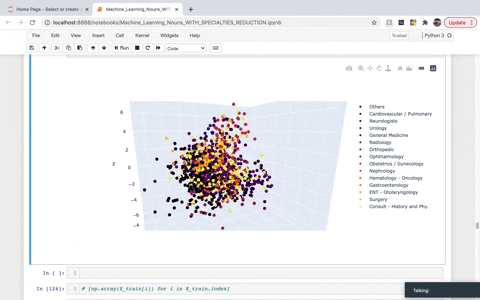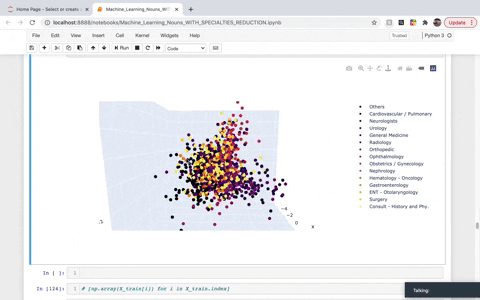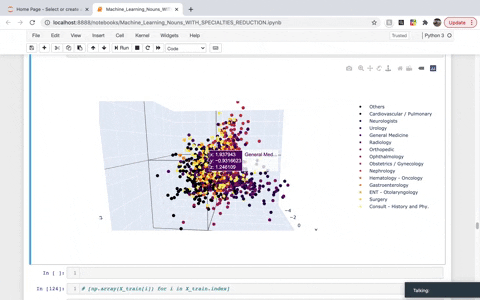
While frequency, one-hot, and TF–IDF encoding enable us to put documents into vector space, it is often useful to also encode the similarities between documents in the context of that same vector space. A word embedding is an approach to provide a dense vector representation of words that capture something about their meaning. To achieve that, we created a list of TaggedDocument objects and then instantiated a Doc2Vec model. We then used Principal Component Analysis (PCA) to reduce the dimensionality to visualize the top three demensions in 3D. The dimensions we are left with in the visual account for roughly 30% of the explained variance.
- Preprocessing the data: cleaning, lemmatizing, POS tagging, tokenizing
- Making the dataset more balanced: finding the right amount to remove/keep/adjust
- Finding the algorithm to use
- Heuristic approach
Why isn't our model performing even better?
-
Medical data in general is a lot harder to analyze / preprocess as it is very complicated
-
In medical transcriptions, there is an overlap in the words that are used. For example, one of our datapoints predicted gastroenterology, however the actual specialty was surgery (could've been surgery of the stomach, so keywords would've overlapped in this example)
-
In clinical notes the same text could be repeated by the same practitioner (a lot of copy and paste)
-
Some medical stopwords could have been removed ("patient", "doctor", "diagnosis") due to a high/high relationship TF/IDF
-
-
We had to make sure to not steer our model and overfit it to show our biases. If we brought too much bias into the process, we would've been taking away the advatange of machine learning.
-
Lack of medical data information
What would make our models better?
- Short term: Random Search Cross Validation using RandomizedSearchCV method: we can define a grid of hyperparameter ranges and randomly sample from the grid. On each iteration, the algorithm will choose a different combination of the features. However, the benefit of a random search is that we are not trying every combination, but selecting at random to sample a wide range of values. Random search will allow us to narrow down the range for each hyperparameter. After that, we will know where to concentrate our search and we will be able to explicitly specify every combination of settings to try. We can do this with GridSearchCV.
- Short term: Customized stopwords vocabulary
- Deep Learning Algorithm
- Spending more time analyzing / cleaning text data (would need subject matter expertise)
- Balancing of the dataset
- Finding more data / medical transcriptions for the specialties with less datapoints