| Branch | Status |
|---|---|
master |
|
develop |
SyntenyViz is a R package to visualise synteny across various biological species.
Visualising the synteny across species not only enables intuitive examination and facilitates reconstruction effort of ancestral genomes, but also allow more direct interrogation of gene regulations and gene structures within a gene cluster.
- Install and load
devtools
install.packages("devtools")
library(devtools)
- Install and load
SyntenyVizfromGitHub
install_github("DPP4ResearchGroup/SyntenyViz")
library(SyntenyViz)
To allow build vignettes, build_vignettes = TRUE options can be used as
install_github("DPP4ResearchGroup/SyntenyViz", build_vignettes = TRUE)
library(SyntenyViz)
Developing version can be accessed via develop as
install_github("DPP4ResearchGroup/SyntenyViz", ref = "develop")
library(SyntenyViz)
Quick and minimum steps to get start a synteney conservation anaysis with SyntenyViz
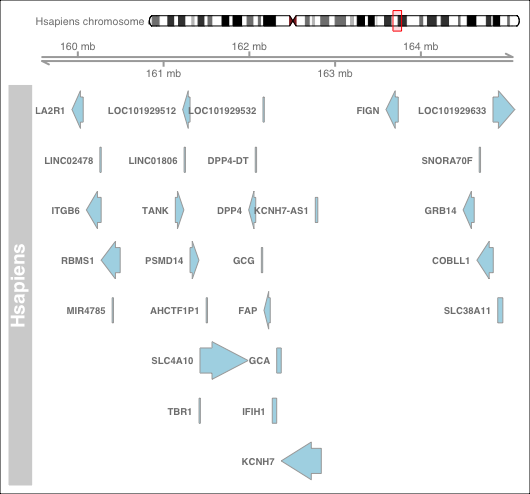
- Define an investigation range We need to firstly define an investigation range to cover the target range in gene coordinate. We will use a mouse dipeptidyl dipeptidase 4 gene (DPP4-mm) in this example, where DPP4-mm locates at chromosome number 2 between 62,330,073-62,412,231 bp.
# orgm is a handle for organism
orgmName <- "Mmusculus"
# mycoords.list is the investigation range handler
mycoords <- "2:6.0e7:6.5e7"
- Convert
mycoords.listinto a GRange object
mycoords.gr <- SyntenyViz::coordFormat (mycoords.list = mycoords)
It is always a good habit to double check the input, so
mycoords.gr
- Construct a single synteny graph
synvizPlot(mycoords.gr, orgmName)
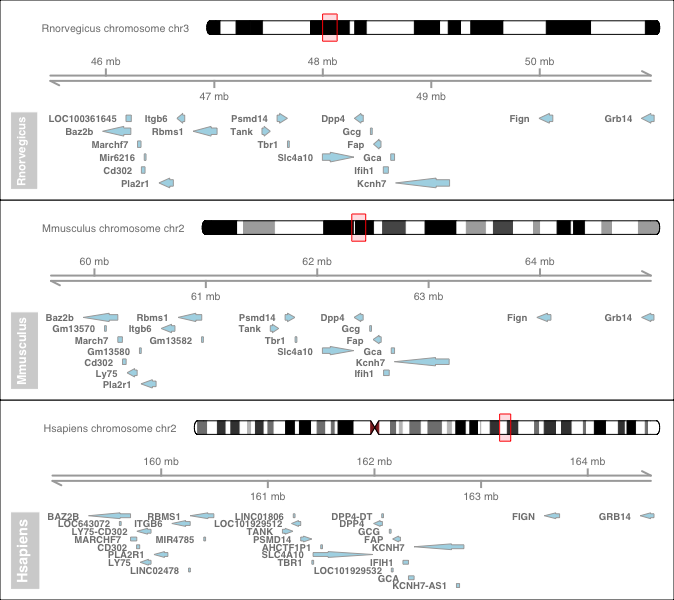
- Construct a multi synteny graph
Pick a few of targets
orgm.1 <- "Hsapiens"
mycoords.list.1 <- "2:15.95e7:16.45e7"
orgm.2 <- "Mmusculus"
mycoords.list.2 <- "2:6.0e7:6.5e7"
orgm.3 <- "Rnorvegicus"
mycoords.list.3 <- "3:4.6e7:5.1e7"
Then construct a multiple synteny query
orgmsList <- orgmsCollection.init (orgmsList)
orgmsList <- orgmsAdd (orgm.1, orgmTxDB, mycoords.list.1, orgmsList)
orgmsList <- orgmsAdd (orgm.2, orgmTxDB, mycoords.list.2, orgmsList)
orgmsList <- orgmsAdd (orgm.3, orgmTxDB, mycoords.list.3, orgmsList)
Now, construct a comparative multi-synteny graph
multiplot <- multisynvizPlots(orgmsList)
Note: Due to heavy downloading and computing involved in this step, multisynvizPlots may take several minutes to complete.
SyntenyViz also includes additional examples and training materials, which can be accessed via vignettes from RStudio
install_github("DPP4ResearchGroup/SyntenyViz", build_vignettes = TRUE)
browseVignettes("SyntenyViz")
OR a PDF can be accessed from SyntenyViz homepage.
- Fork to your contributing account
- Create your feature branch (
git checkout -b my-new-feature) - Commit your changes (
git commit -am 'Added some feature') - Push to the feature branch (
git push origin my-new-feature) - Create a new PR
Issues and bugs can be raised and tracked through GitHub issue tracker for SyntenyViz.
Travis CI testing (travis status) inplements R CMD check.
The function integrity is checked by R native testthat, which can also be invoked by utility function devtools::test() from RStudio.