No longer maintained ... please check out blobtools
Light version of the upcoming blobtools package
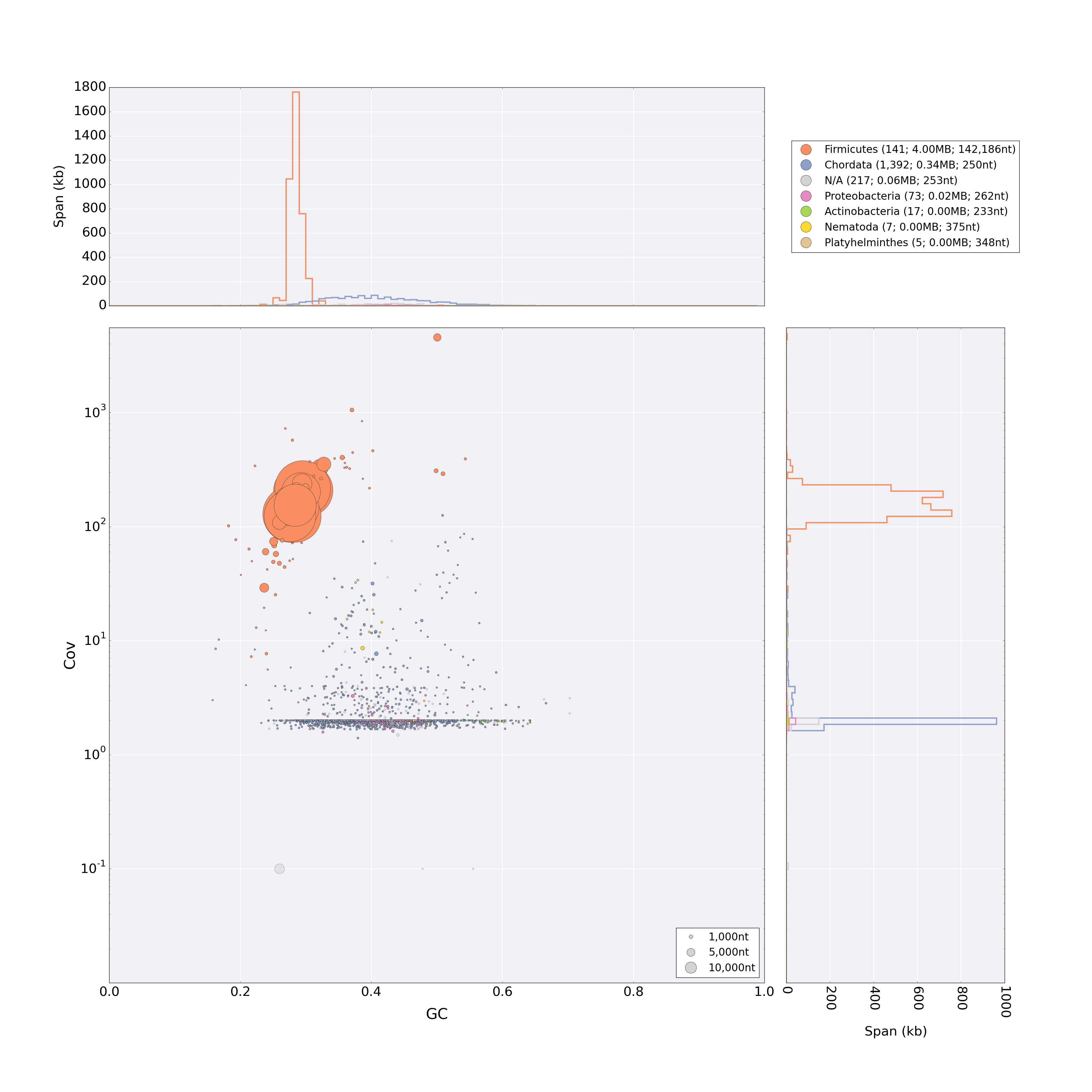
blobtools-light allows the visualisation of (draft) genome assemblies using TAGC (Taxon-annotated Gc-Coverage) plots (Kumar et al. 2012).
Software requirements:
- Python 2.7+
- Numpy 1.9.1 ('pip install numpy')
- matplotlib 1.4 ('pip install matplotlib')
- NCBI Taxdump (' wget ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/taxdump.tar.gz ')
Data requirements:
- Assembly* (FASTA)
- if assembly was generated through Spades, Abyss or Velvet, coverage can be parsed from contig headers (use e.g. -spades SPADESASSEMBLY); if coverage from assembly is not needed -exclude_assembly_cov can be specified
- Coverage information (one or more):
- COV (Tab-separated: "header\tcoverage")
- BAM/SAM**
- CAS format** (CLC mapper) ** The first time BAM/SAM/CAS file(s) gets parsed, the program will create a COV file for each. If omeanalsysis
- BLAST result (using BLAST 2.2.29+):
- suggested command : "blastn -task megablast -query ASSEMBLY.fa -db /blast_db/nt -outfmt '6 qseqid staxids bitscore std sscinames sskingdoms stitle' -max_target_seqs 25 -culling_limit 2 -num_threads 16 -evalue 1e-25 -out ASSEMBLY.vs.nt.25cul1.1e25.megablast.out"
- The important part is "-outfmt '6 qseqid staxids bitscore'"
Making blobplot file:
usage: makeblobs.py -a <ASSEMBLY> -cas <CAS> -blast <BLAST> -taxdb <PATH_TO_TAXDB> -o <OUTPUT> [-h]
optional arguments:
-h, --help show this help message and exit
-a ASSEMBLY_FASTA Assembly file
-spades SPADES_FASTA SPADES assembly file
-velvet VELVET_FASTA VELVET assembly file
-abyss ABYSS_FASTA ABYSS assembly file
-exclude_assembly_cov Exclude coverage from assembly file
-cov COV_FILE [COV_FILE ...] COV (mapping) file(s)
-bam BAM_FILE [BAM_FILE ...] BAM (mapping) file(s)
-sam SAM_FILE [SAM_FILE ...] SAM (mapping) file(s)
-cas CAS_FILE [CAS_FILE ...] CAS (mapping) file(s)
-blast BLAST_FILE [BLAST_FILE ...] BLAST file(s)
-rank TAX_RANK Select target taxonomic rank (species, genus, order,
phylum, superkingdom). Default = phylum
-taxrule A or B Tax-rule on how to deal with multiple BLAST libs. A :
"higher bitscore wins", B : "Decreasing trust in BLAST
libs"
-taxdb TAX_DUMP Path to NCBI taxdb (nodes.dmp, names.dmp)
-o OUTPUT_PREFIX Output prefix
-v show program's version number and exit
>> ~/blobtools-light/makeblobs.py -a assembly/assembly.fa -bam mapping/assembly.mapping.bam -taxdb /exports/blast_db/ -blast assembly.vs.nt.25cul1.1e25.megablast.out -o test
Plotting blobplot-file:
~/blobtools-light/plotblobs.py test.blobplot.txt