Introduction
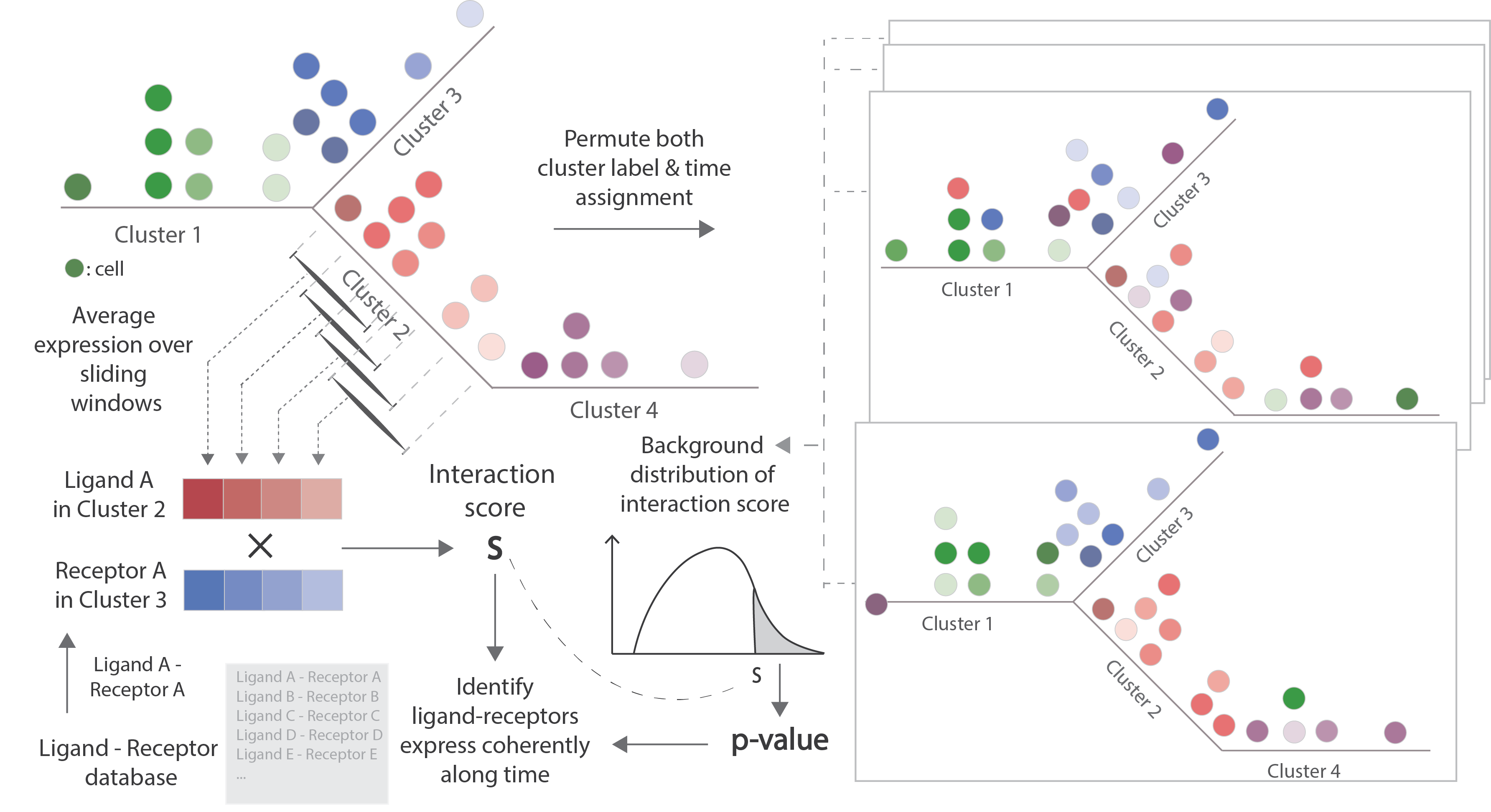
TraSig (Trajectory-based Signalling genes inference) identifies interacting cell types pairs and significant ligand-receptors based on the expression of genes as well as the pseudo-time ordering of cells. For any two groups of cells that are expected to overlap in time, TraSig takes the pseudo-time ordering for each group and the expression of genes along the trajectory as input and then outputs an interaction score and p-value for each possible ligand-receptor pair. It also outputs a summary score for cell type pairs by combining individual ligand-receptors' scores.
Table of Contents
Get-started
Prerequisites
- Python >= 3.6
- Python side-packages:
-- numpy >= 1.19.5
-- pandas >= 0.23.4
-- Bottleneck >= 1.3.2
-- statsmodels >= 0.12.1 (required for post-analysis only)
-- scipy >= 1.5.4 (required for post-analysis only)
-- matplotlib >= 3.3.4 (required for post-analysis only)
-- seaborn >= 0.11.0 (required for post-analysis only)
Installation
Install within a virtual environment
It is recommended to use a virtural environment/pacakges manager such as Anaconda. After successfully installing Anaconda/Miniconda, create an environment by following:
conda create -n myenv python=3.6You can then install and run the package in the virtual environment. Activate the virtural environment by:
conda activate myenvMake sure you have pip installed in your environment. You may check by
conda listIf not installed, then:
conda install pipThen install TraSig, together with all its dependencies by:
pip install git+git://github.com/doraadong/TraSig.gitIf you want to upgrade TraSig to the newest version, then first uninstall it by:
pip uninstall trasigAnd then just run the pip install command again.
Not using virtural environment
If you prefer not to use a virtual envrionment, then you may install TraSig and its dependencies by (may need to use sudo):
pip3 install git+https://github.com/doraadong/TraSig.gitYou may find where the package is installed by:
pip show trasigCommand-line
Run TraSig
Run TraSig by (arguments are taken for example):
main.py -i input -o output -d oligodendrocyte-differentiation-clusters_marques -g None -b ti_slingshot -n 1000 -s smallerWindowThe usage of this command is listed as follows:
usage: main.py [-h] -i INPUT -o OUTPUT -d PROJECT -g PREPROCESS -b MODELNAME
[-t LISTTYPE] [-l NLAP] [-m METRIC] [-z NAN2ZERO] [-n NUMPERMS]
[-p MULTIPROCESS] [-c NCORES] [-s STARTINGTREATMENT]
[-a ALIGNTYPE] [-y GENEPAIRTYPE] [-f SMOOTH] [-v OVERLAP]
[-r RATE] [-e ERRORTYPE] [-k ARATE] [-j BRATE]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
string, folder to find inputs
-o OUTPUT, --output OUTPUT
string, folder to put outputs
-d PROJECT, --project PROJECT
string, project name
-g PREPROCESS, --preprocess PREPROCESS
string, preprocessing steps applied to the data /
project, default None
-b MODELNAME, --modelName MODELNAME
string, name of the trajectory model
-t LISTTYPE, --listType LISTTYPE
string, optional, interaction list type, default
ligand_receptor
-l NLAP, --nLap NLAP integer, optional, sliding window size, default 20
-m METRIC, --metric METRIC
string, optional, scoring metric, default dot
-z NAN2ZERO, --nan2zero NAN2ZERO
boolean, optional, if treat nan as zero, default True
-n NUMPERMS, --numPerms NUMPERMS
integer, optional, number of permutations, default
10000
-p MULTIPROCESS, --multiProcess MULTIPROCESS
boolean, optional, if use multi-processing, default
True
-c NCORES, --ncores NCORES
integer, optional, number of cores to use for multi-
processing, default 4
-s STARTINGTREATMENT, --startingTreatment STARTINGTREATMENT
string, optional, way to treat values at the beginning
of an edge with sliding window size smaller than nLap,
None/parent/discard/smallerWindow, default
smallerWindow, need to provide an extra input
'path_info.pickle' for 'parent' option
-a ALIGNTYPE, --alignType ALIGNTYPE
string, optional, how to align edges, options:
unaligned/aligned-fixed/aligned-specific, default
unaligned
-y GENEPAIRTYPE, --genePairType GENEPAIRTYPE
string, optional, identifier for the type of genes to
align, e.g. interaction/cell_cycle, default
interaction
-f SMOOTH, --smooth SMOOTH
float, optional, smoothing parameter for splines,
default 1
-v OVERLAP, --overlap OVERLAP
float, optional, overlap threshold for alignment,
default 0.5
-r RATE, --rate RATE integer, optional, sampling rate for aligned time
points, default 1
-e ERRORTYPE, --errorType ERRORTYPE
string, optional, type of distance metric for
alignment (MSE, cosine or corr), default cosine
-k ARATE, --aRate ARATE
float, optional, rate to sample parameter a for
alignment, default 0.05
-j BRATE, --bRate BRATE
float, optional, rate to sample parameter b for
alignment, default 2.5Prepare inputs for TraSig (from dynverse outputs)
For preparing inputs using user-defined trajectory (not from dynverse), see tutorial.
Given dynverse outputs, prepare inputs by (arguments are taken for example):
python prepare_inputs.py -i ../trajectory/input -o ../example/input -d oligodendrocyte-differentiation-clusters_marques -t ../trajectory/output/output.h5 -g None -b ti_slingshot -e NoneThe usage of this command is listed as follows:
usage: prepare_inputs.py [-h] -i INPUT -o OUTPUT -d PROJECT -t TRAJECTORYFILE
-g PREPROCESS -b MODELNAME [-e OTHERIDENTIFIER]
[-c LISTTYPE] [-cp PATHLR] [-y GENEPAIRTYPE]
[-yp PATHALIGN]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
string, folder to find inputs for trajectory inference
-o OUTPUT, --output OUTPUT
string, folder to save inputs for TraSig
-d PROJECT, --project PROJECT
string, project name
-t TRAJECTORYFILE, --trajectoryFile TRAJECTORYFILE
string, trajectory output file from dynverse, default
../trajectory/output/output.h5
-g PREPROCESS, --preprocess PREPROCESS
string, preprocessing steps applied to the data /
project, default None
-b MODELNAME, --modelName MODELNAME
string, name of the trajectory model
-e OTHERIDENTIFIER, --otherIdentifier OTHERIDENTIFIER
string, optional, other identifier for the output,
default None
-c LISTTYPE, --listType LISTTYPE
string, optional, interaction list type, default
ligand_receptor
-cp PATHLR, --pathLR PATHLR
string, optional, path to the interaction list,
default
../ligand_receptor_lists/ligand_receptor_FANTOM.pickle
-y GENEPAIRTYPE, --genePairType GENEPAIRTYPE
string, optional, identifier for the type of genes to
align, e.g. interaction/cell_cycle, default
interaction
-yp PATHALIGN, --pathAlign PATHALIGN
string, optional, path to the alignment genes list,
set as 'None' if not doing alignment or using
'interaction' for alignment, default None
Analyze outputs from TraSig
Analyze outputs by (arguments are taken for example):
python analyze_outputs.py -i ../example/input -o ../example/output -d oligodendrocyte-differentiation-clusters_marques -g None -p None -b ti_slingshot -p None -n 100000 -s smallerWindowThe usage of this command is listed as follows:
usage: analyze_outputs.py [-h] -i INPUT -o OUTPUT -d PROJECT -g PREPROCESS -b
MODELNAME [-t LISTTYPE] [-p OTHERIDENTIFIER]
[-l NLAP] [-m METRIC] [-z NAN2ZERO] [-n NUMPERMS]
[-s STARTINGTREATMENT] [-a ALIGNTYPE]
[-y GENEPAIRTYPE] [-f SMOOTH] [-v OVERLAP] [-r RATE]
[-e ERRORTYPE] [-k ARATE] [-j BRATE]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
string, folder to find TraSig's inputs
-o OUTPUT, --output OUTPUT
string, folder to find TraSig's outputs
-d PROJECT, --project PROJECT
string, project name
-g PREPROCESS, --preprocess PREPROCESS
string, preprocessing steps applied to the data /
project, default None
-b MODELNAME, --modelName MODELNAME
string, name of the trajectory model
-t LISTTYPE, --listType LISTTYPE
string, optional, interaction list type, default
ligand_receptor
-p OTHERIDENTIFIER, --otherIdentifier OTHERIDENTIFIER
string, optional, other identifier for the output,
default None
-l NLAP, --nLap NLAP integer, optional, sliding window size, default 20
-m METRIC, --metric METRIC
string, optional, scoring metric, default dot
-z NAN2ZERO, --nan2zero NAN2ZERO
boolean, optional, if treat nan as zero, default True
-n NUMPERMS, --numPerms NUMPERMS
integer, optional, number of permutations, default
10000
-s STARTINGTREATMENT, --startingTreatment STARTINGTREATMENT
string, optional, way to treat values at the beginning
of an edge with sliding window size smaller than nLap,
None/parent/discard/smallerWindow, default
smallerWindow, need to provide an extra input
'path_info.pickle' for 'parent' option
-a ALIGNTYPE, --alignType ALIGNTYPE
string, optional, how to align edges, options:
unaligned/aligned-fixed/aligned-specific, default
unaligned
-y GENEPAIRTYPE, --genePairType GENEPAIRTYPE
string, optional, identifier for the type of genes to
align, e.g. interaction/cell_cycle, default
interaction
-f SMOOTH, --smooth SMOOTH
float, optional, smoothing parameter for splines,
default 1
-v OVERLAP, --overlap OVERLAP
float, optional, overlap threshold for alignment,
default 0.5
-r RATE, --rate RATE integer, optional, sampling rate for aligned time
points, default 1
-e ERRORTYPE, --errorType ERRORTYPE
string, optional, type of distance metric for
alignment (MSE, cosine or corr), default cosine
-k ARATE, --aRate ARATE
float, optional, rate to sample parameter a for
alignment, default 0.05
-j BRATE, --bRate BRATE
float, optional, rate to sample parameter b for
alignment, default 2.5
Tutorials
Github rendering disables some functionalities of Jupyter notebooks. We recommend using nbviewer to view the following tutorials.
Run TraSig on example data and analyze outputs
The example inputs and outputs can be found under the folder example. You may follow the tutorial to run TraSig on the example data and analyze the outputs. You may also obtain the analysis outputs by running the aforementioned script analyze_outputs using command-line. See the tutorial for more details.
Prepare inputs
To run TraSig, we need to have 4 input files. Here is a tutorial, showing how to prepare these files from the inference results of any trajectory inference method included in dynverse. You can find the example expression data (input) and trajectory inference result (output) under the folder trajectory. You may also prepare the inputs for TraSig by running the aforementioned script prepare_inputs using command-line. See the tutorial for more details.
We can also accept inputs that are not generated by dynverse. For outputs from any pseudotime trajectory tool you prefer, you can prepare the inputs for TraSig following this tutorial.
We can also accept customized ligand-receptor database and customized gene list for alignment, if you would like to use the alignment option for TraSig. The inputs will be changed accordingly and you may need to specify the filepath and identifier for your own ligand-receptor database and gene list for alignment. Please find the corresponding arguements in the command-line tool and the corresponding variables in the tutorials mentioned above to make the changes. You may also find descriptions on the file formats in the tutorials. The example ligand-receptor database is from [1] and the example alignment gene lists is downloaded from the Seurat package.
Updates-log
-
2-2-2022:
-- Add support for conducting temporal alignment using customized gene list -
12-21-2021:
-- Add support for conducting temporal alignment and calculating scores using optimally aligned expression profiles
-- Add tutorial illustrating how to prepare inputs using user-defined trajectory, not necessarily from dynverse
Learn-more
Check our preprint at biorxiv.
Credits
The software is an implementation of the method TraSig, jointly developed by Dora Li, Jun Ding and Ziv Bar-Joseph from System Biology Group @ Carnegie Mellon University. We also acknowledge Jeremy J. Velazquez, Joshua Hislop and Mo R. Ebrahimkhani from University of Pittsburgh for the fruitful discussions on method development.
Contacts
- dongshul at andrew.cmu.edu
License
This project is licensed under the MIT License - see the LICENSE file for details
References
[1] Ramilowski, J., Goldberg, T., Harshbarger, J. et al. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat Commun 6, 7866 (2015). https://doi.org/10.1038/ncomms8866