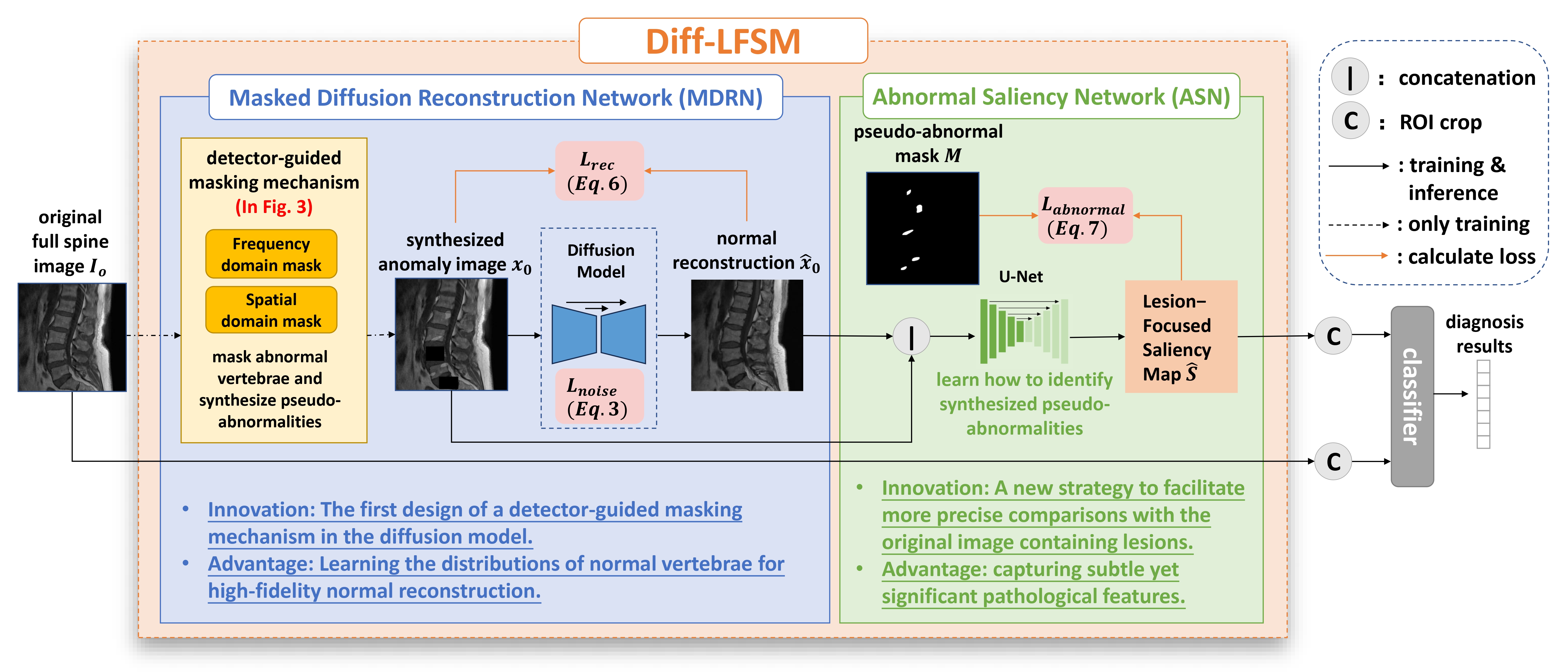
Learning Interpretable Lesion-Focused Saliency Maps with Context-Aware Diffusion Model for Vertebral Disease Diagnosis
To set up the environment, use the requirements.txt file to prepare the Conda environment. Ensure you have all necessary dependencies installed.
conda env create -f environment.yml
conda activate diffcasm
As an example, to prepare the VerTumor600 dataset, convert the MRI vertebrae data from JSON to PNG format using the provided script:
python ./data/VerTumor600/MRI_vertebrae/json_to_png.py
To train the Diff-CASM model, run the following command:
python diff_training_seg_training.py ARG_NUM=1
Once the Diff-CASM model is trained, use it to generate the context-aware saliency maps (CASM):
python diff_training_seg_training.py ARG_NUM=2
After generating CASMs, crop out individual vertebrae from the images. For the VerTumor600 dataset, use the script below:
python ./data/VerTumor600/cropped_vertebrae/crop_images.py
python spine_trans_cls_with_FE.py ARG_NUM=3
Thanks to the following works: AnoDDPM, guided-diffusion.