| description |
|---|
OTU table visualization, data analysis, and machine learning |
Mian is an open-source web-platform that enables data exploration and analysis of OTU or ASV data with respect to gene expression, categorical and numerical metadata (eg. immunohistological data, experimental conditions), and taxonomic structure.
Mian gives researchers access to interactive data visualizations, machine learning models and feature selection tools, and automated statistical assessments. No code required.
- Apply linear regression or a deep-neural network on your data without any additional setup
- Automatically find important genes or histological features
- Track your work over time with experiment notebooks
- Bucket quantitative measures to access categorical tools (eg. define thresholds to categorize different severities of a disease)
- Filter by specific taxonomic groups, OTUs, or sample groups
Common features include detail highlights, tuneable settings, filters, notebook snapshots, and downloadable images.
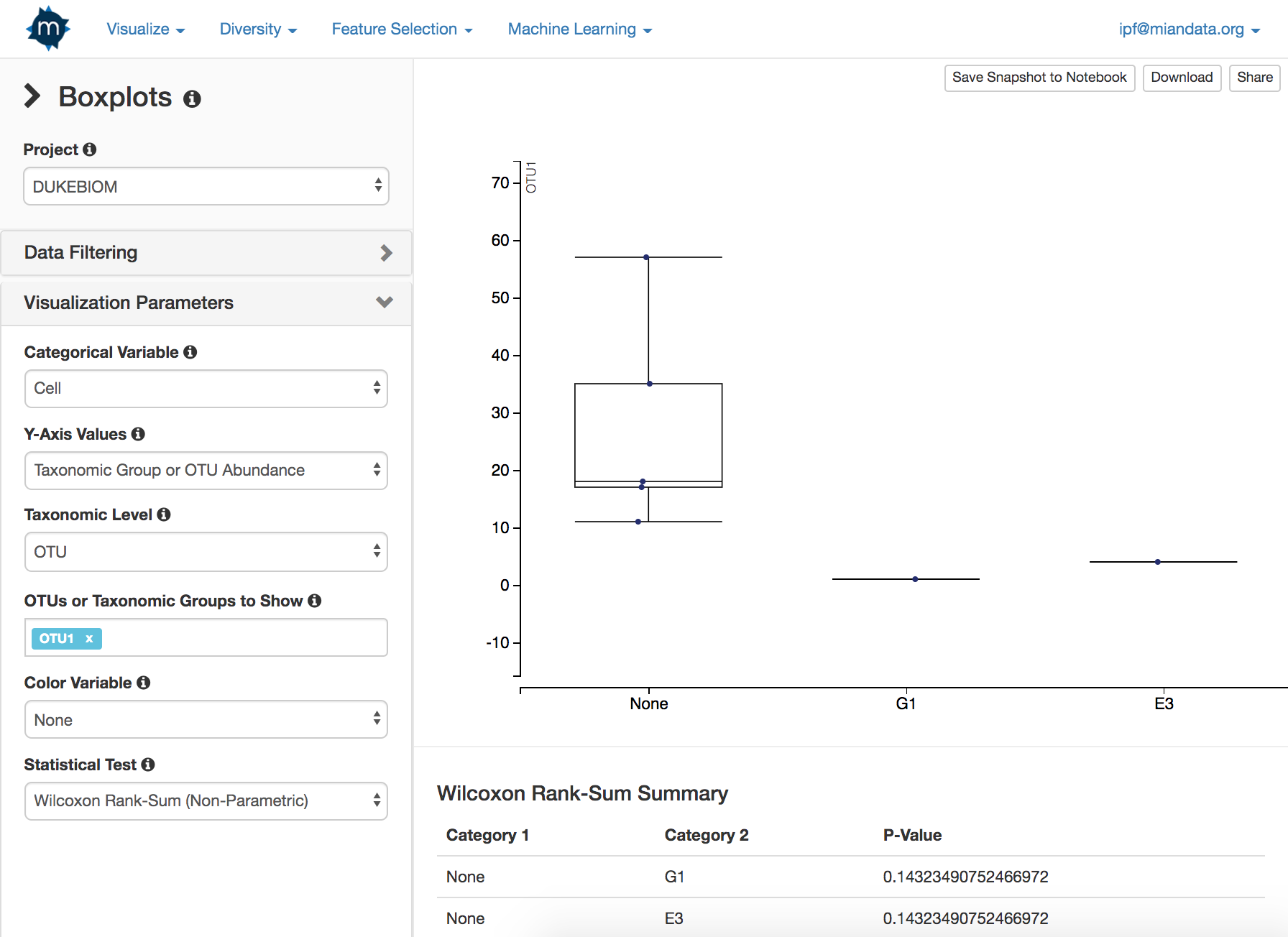
- Boxplots
- Create boxplots of specific OTUs or taxonomic groups against categorical variables
- Apply color according to a second categorical variable
- Automated parametric/non-parametric statistical tests
- Barplots or Donut Charts
- Examine OTU composition at different taxonomic levels or categorical variables
- Scatterplots
- Plot OTU composition, gene expression, alpha diversity, or aggregate counts against each other
- Color by categorical variables or resize points by quantitative variables
- Heatmap
- Explore correlations of OTU counts or quantitative variables
- Visualize composition of taxonomic groups across samples
- Correlation Network
- Cluster correlated samples and color by categorical variables
- Rarefaction Curves
- Subsampling depths at varying sample sizes
- Taxonomic Trees
- Compare OTU counts on a taxonomic tree across different categorical variables
Explore different microbial diversity measures using boxplots, scatterplots, and NMDS/PCoA plots.
- Alpha Diversity
- Compare species diversity measures on a boxplot or scatterplot across sample groups
- Available measures include: Shannon, Simpson, Faith's Phylogenetic Diversity
- Beta Diversity
- Compare differences of diversity between different sample groups using a boxplot
- Available measures include: Bray-Curtis, Jaccard, Sorenson, Whittaker, Weighted/Unweighted Unifrac
- NMDS plot
- Visualize beta diversity using a non-metric multidimensional scaling plot on different sample groups
- PCoA plot
- 2D or 3D principal component analysis using different distance measures and categorical variables
Automatically find OTUs, taxonomic groups, or metadata that can either correlate well together, or were determined to be important due to a statistical test.
- Boruta (Random Forest) Classification
- Find the OTUs or taxonomic groups that differentiate sample groups according to a Random Forest using the Boruta feature selection algorithm
- Elastic Net Classification and Regression
- Use varying regularization in Elastic Net to find OTUs or taxonomic groups that differentiate between sample groups
- Assess important of selected features using a machine learning model
- Fisher Exact Test
- Apply a presence-absence test on OTUs or taxonomic groups across different sample groups
- Differential Selection
- Look for OTUs or taxonomic groups that differentiate between two sample groups according to a statistical test and corresponding FDR-corrected q-value
- Correlation Selection
- Look for genes, OTUs or taxonomic groups, or quantitative metadata that correlate with another gene , OTU or taxonomic group, quantitative metadata, or alpha diversity according to a statistical test and corresponding FDR-corrected q-value
Assess the ability of your dataset to generalize using a machine learning model by creating a classifier or regressor on a training dataset and evaluating on a test dataset.
- Linear Regressor and Classifier
- Choose an experimental variable to train and test the ability of your linear model (with regularization) to predict the variable
- Random Forest Classifier
- Choose an categorical variable to train and test the ability of a random forest model to predict the variable
- Deep Learning
- Design a deep neural network with dropout to assess the ability of the model to predict a chosen variable
Mian is built using HTML/CSS/JavaScript for the front-end and Python and R for the back-end.
- Bootstrap 3: CSS styling
- jQuery: Front-end dynamic website interaction handling
- Flask: Python-based back-end server
- flask-login: Enables login capabilities for the Mian website
- biom-format: Allows processing of user-uploaded standard biological matrix format (BIOM)
- h5py: Allows processing of HDF5 formatted files
- rpy2: Interface between Python and R code
- scikit-learn: Machine learning algorithms implemented in Python
- scipy: Python library used for scientific processing
- werkzeug: WSGI application for Python
- scikit-bio: Python library used for bioinformatics
- pandas: Provides high-performance data structure and matrix manipulation
- vegan: R library for community ecology analysis
- RColorBrewer: Color maps for R
- ranger: Fast random forest implementation
- Boruta: All-relevant feature selection algorithm
- glmnet: Lasso and elastic-net regularized linear models
Mian is currently described in a pre-print paper in bioRxiv: https://www.biorxiv.org/content/early/2018/09/14/416073
Mian is built at the University of British Columbia and supported by the Providence Health Care Research Institute and Centre for Heart Lung Innovation at St. Paul's Hospital