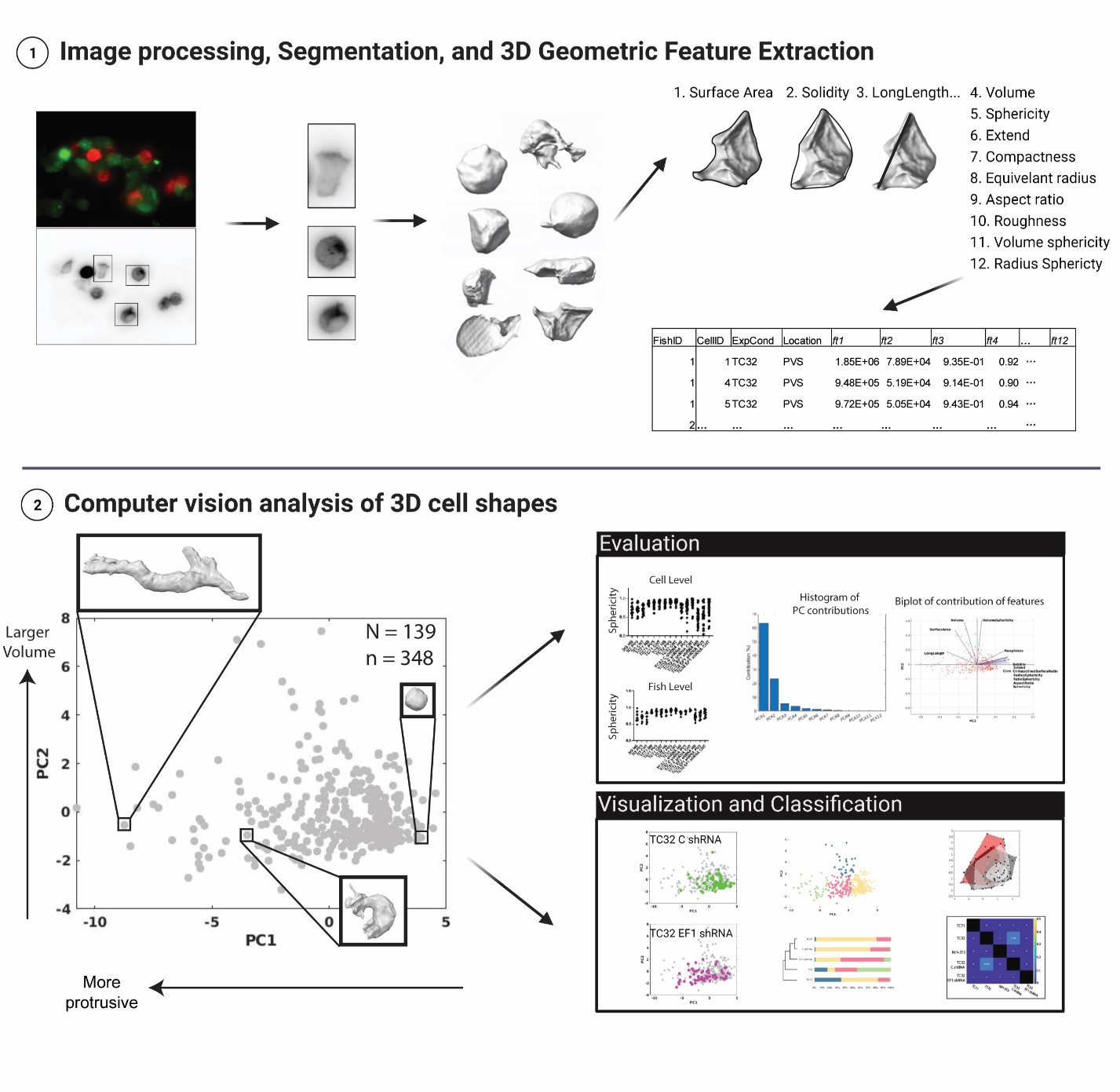
3dCellShape implements measurement and analysis of geometric features on extracted 3D volumes of single cells from fluorescent imaging data.
- This repository is for initial distribution of the 3D shape analysis pipeline associated with the manuscript In vivo 3D profiling of site-specific human cancer cell morphotypes in zebrafish, JCB, 2022, 221 (11): e202109100, written by Dagan Segal, Hanieh Mazloom-Farsibaf, Bo-Jui Chang, Philippe Roudot, Divya Rajendran, Stephan Daetwyler, Reto Fiolka, Mikako Warren, James F. Amatruda, Gaudenz Danuser.
- The pipeline consists of three Matlab scripts – (1) for post-processing and (2) segmentation of data, adapted from Driscoll, et al, 2019, and (3) for geometric feature extraction and analysis from extracted 3D volumes.
- Example input data is provided in the link exampleCroppedCells. These are cropped regions of interest from raw data containing a single fluorescently labeled cell. Using cropped regions rather than entire raw data files as input enables a more time-efficient data processing, as the file sizes are significantly smaller. The folder also includes an example “index” file detailing the relevant parameters of the experiment. For proper segmentation, the fluorescently labeled cell must not be in contact with any other cell or image boundary, in order to properly capture cell boundaries.
- Corresponding example output files are provided in the link exampleOutput.
- Workflow:
- i. (Part 1) Run image registration and deconvolution of cropped raw data by running imageProcessing3D.m in the PostProcessing_pipeline folder. This step is optional.
- ii. (Part 2) and segmentation of cell volumes by running runMorphology3D.m. The subfunctions for this script are included in the Segmentation_pipeline folder.
- iii. (Part 3) Run runGlobalFeatureAnalysis.m to measure geometric features of cells from 3D segmented data, create an indexed table of geometric features, and create some basic visualizations of data. This script is found under the Global_Feature_Analysis folder.
- Pipeline built on MATLAB R2020a version