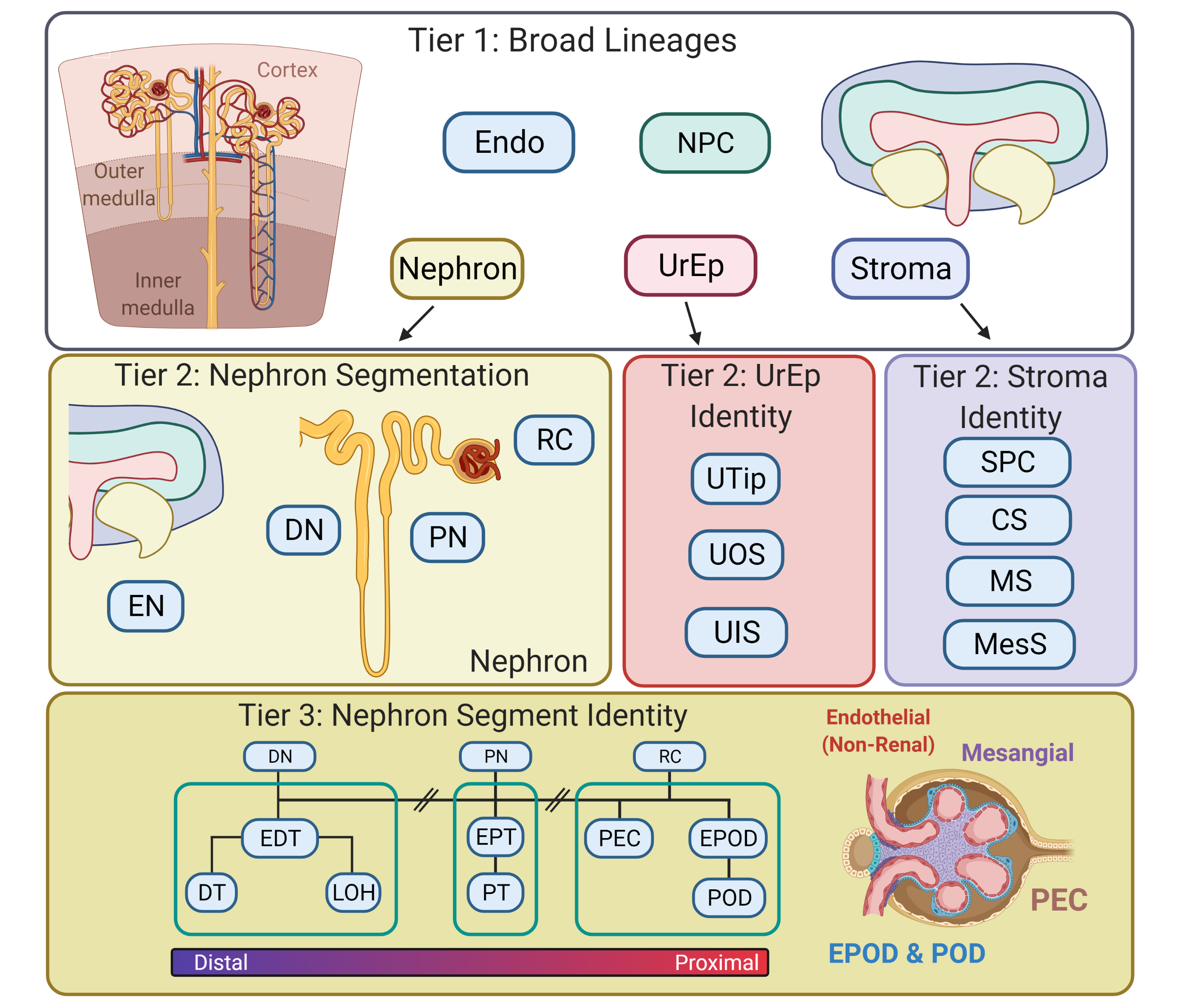
DevKidCC (Developing Kidney Cell Classifier) is a tool that will classify single cell kidney data, both human tissue and human stem cell derived organoids. There is no pre-processing required, although we do recommend filtering out poor quality cells for most accurate representation of cell proportions.
bioRxiv paper: Wilson et al. 2021
You can install DevKidCC from this repository using devtools:
# prerequite scPred package
devtools::install_github("powellgenomicslab/scPred")
# DevKidCC itself
devtools::install_github("KidneyRegeneration/DevKidCC", ref = "main")Expected installation time is under 5 minutes for each package.
This package has been successfully tested on both Windows and Linux systems.
Check out the full vignette which includes details on using the visualisation functions here
DevKidCC operates on single cell data as a Seurat object. The simplest workflow to use DevKidCC is:
library(DevKidCC)
# read in seurat object
organoid <- DKCC(organoid) # use dataset 'organoid' included in packageThis will cause a number of additional metadata to be added to the object. The first tier is labelled "LineageID" while the complete annotation is under "DKCC"
The database is stored as an rda file and can be downloaded at the following link: https://drive.google.com/file/d/1wh551HvecgszizE8FCsXRD5CiQVYm3K6/view?usp=sharing
Save this file in a data folder at the top level of your project. This will allow it to be loaded by the DotPlotCompare function when compare.to.organoids = T
Any issues submit a pull request or contact me at sean.wilson@mcri.edu.au