user@machine:~$ mkdir ecosystem; cd ecosystem
user@machine:~$ git clone https://gitlab.pnnl.gov/dadaist/neuromancer.git
user@machine:~$ git clone https://gitlab.pnnl.gov/dadaist/psl.git
user@machine:~$ git clone https://gitlab.pnnl.gov/dadaist/slim.git
# Resulting file structure:
ecosystem/
neuromancer/
psl/
slim/user@machine:~$ conda env create -f env.yml
(neuromancer) user@machine:~$ source activate neuromanceruser@machine:~$ conda config --add channels conda-forge pytorch
user@machine:~$ conda create -n neuromancer python=3.7
user@machine:~$ source activate neuromancer
(neuromancer) user@machine:~$ conda install pytorch torchvision -c pytorch
(neuromancer) user@machine:~$ conda install scipy pandas matplotlib control pyts numba scikit-learn mlflow dill
(neuromancer) user@machine:~$ conda install -c powerai gym(neuromancer) user@machine:~$ cd psl
(neuromancer) user@machine:~$ python setup.py develop
(neuromancer) user@machine:~$ cd ../slim
(neuromancer) user@machine:~$ python setup.py develop
(neuromancer) user@machine:~$ cd ../neuromancer
(neuromancer) user@machine:~$ python setup.py develop[ ] (Jan & Shri) Confirm environment set up works on Mac and Linux
[ ] (Next coding session) Put generic control script with tutorial system as default (use common.py style refactor)
[ ] (Jan) EED building sys id papers code release: port to neuromancer to recreate experiments
[ ] (Elliott) prepare constrained_block_ssm_acc for release
[ ] (Jan) prepare flexy for release
[ ] (Elliott & Soumya) prepare lpv_14dc for release
[ ] (Aaron) break off neural koopman branch since this code won't be released in the initial release
[ ] (Aaron) Create docs
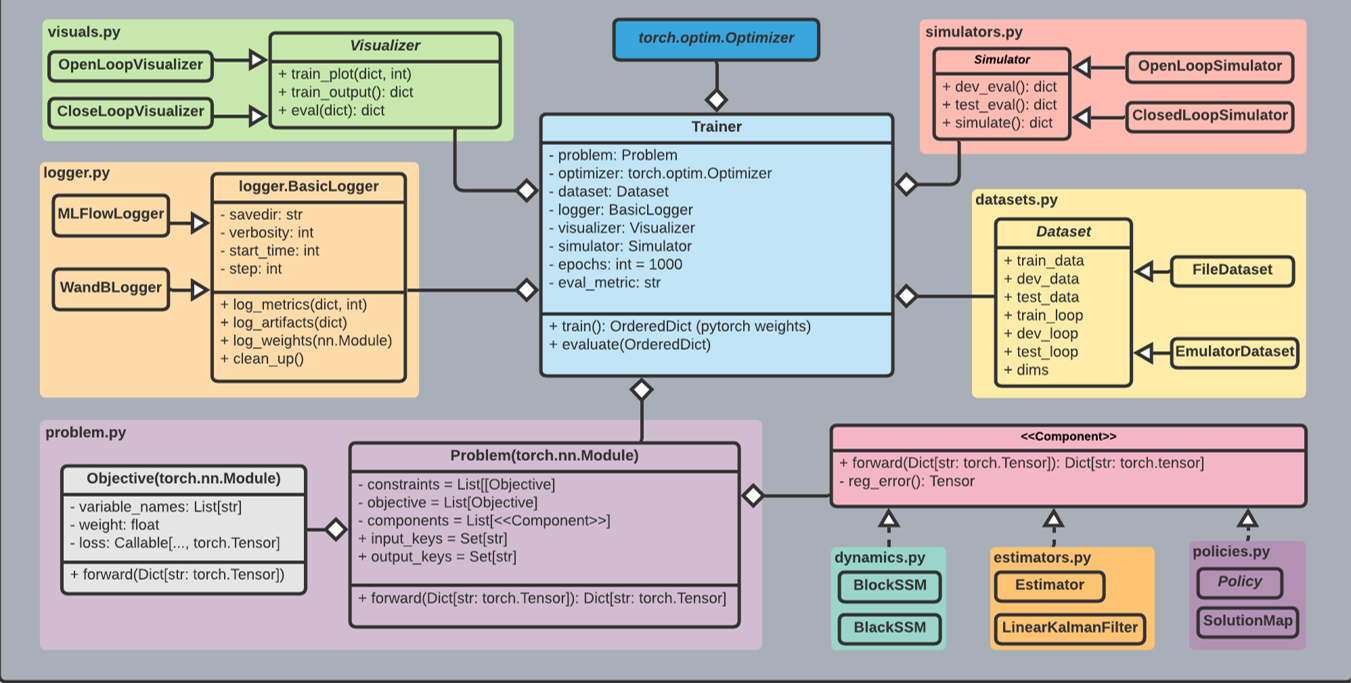
[1] User experience updates
new attributes for our component models:
out_keys - to have a full info about the connectivity
new attributes for models and objectives:
form or equation - a string representing the underlying mathematical expression e.g. "y = x + f(x)"
pre-defined constraints in a separate file with brief documentation
[2] datasets
refactor datasets.py - get rid of unnecessary dependencies
custom normalizations: [-6, 6] etc...
[3] create new analysis file
post hoc analysis of neural nets
decouple eigenvalue analysis from visuals in trainer
add eigenvalue analysis for all weights, neural blocks, and all activation types
[4] trainer
keep current trainer
set random seeds for reproducibility
add profiling
[5] logger
add save weights option during training for running offline visuals and analysis
[6] dynamics, estimator, policies
no immediate actions
reduce code by generalizing model classes and helper functions
[7] documentation
autogenerate docs via doc strings
python package style format with latex syntax
datasets category [ ] In datasets add data_transforms method to act on dataset.data to generate: finite difference sequences via np.diff, nonlinear expansions, spectral decompositions [ ] unify batching/unbatching via single function in datasets.py? [ ] finish batch_data_exp_id for datasets generated via multiple experiment runs: batch based on exp_idx and nsteps [ ] Mini-batching
visuals category [ ] update plot_matrix method in VisualizerOpen(Visualizer) - currently supports only linear maps with effective_W [ ] Visualizer for Multi-parametric programs [ ] Visualize learnable loss function evolution [ ] stream plots for phase spaces of ODEs
problem modeling and training script functions category [ ] move freeze_weight, unfreeze_weight, and share_weights into problem.py? [ ] add output_keys attribute to components [ ] add input_keys and output_keys attributes to overall model generated by Problem() [ ] Generalize sliding window between 1 and nsteps
trainer category [ ] Learn-rate scheduling [ ] WandB logger
blocks, dynamics, policies, estimators category
[ ] Re-implement RNN state preservation for open loop simulation
[ ] full trajectory estimators: This will entail only taking the first N-steps for all the non-static inputs
[ ] Pytorch Extended Kalman Filter:
https://filterpy.readthedocs.io/en/latest/_modules/filterpy/kalman/EKF.html
[ ] Implement LQR policy, similar structure to Linear Kalman Filter:
Scipy reference https://nbviewer.jupyter.org/url/argmin.net/code/little_LQR_demo.ipynb
dissemination and documentation category [ ] Doc strings [ ] Sphinx docs [ ] Package distribution via conda or pypi [ ] Look at this testing software to for automatic wider test coverage: https://hypothesis.readthedocs.io/en/latest/