This is aimed to show the three types transcriptome-wide association study (TWAS) in our publication, and give instructions on how to integrate our soybean SeedlingShoot eQTLs into your own GWAS.
- Prerequisites
- TWAS using measured expression
- TWAS using imputed expression -- FUSION
- TWAS using mendelian randomization -- SMR
- FAQ
- Citation
- Acknowledgments
Requirements to run the analyses:
-
Data The reads counts for gene and coding exon, expression weights required by FUSION, summary level data required by SMR, and pod color TWAS with three methods are available on FigShare.
unzip xxxx.zip #readme.txt gave description on the files. -
TWAS -- Measured Expression
-
Rstudio (R editor, recommended but not required)
-
R packages:
#installation of GAPIT -- solution I source("http://zzlab.net/GAPIT/GAPIT.library.R") source("http://zzlab.net/GAPIT/gapit_functions.txt") #installation of GAPIT -- solution II install.packages("devtools") devtools::install_github("jiabowang/GAPIT3",force=TRUE) library(GAPIT3)
-
TWAS -- SMR by Yang Lab
-
TWAS -- Fusion by Gusev Lab
In a previous repository from an indepent publication (Li, 2021), TWAS with measured expression was described. In the current paper, we extend the measured expression from gene expression to include exon expression and exon proportion (reads ratio of each coding exon in certain gene). By capturing more expression variation, these three types of measured expression allow for a more comprehensive understanding of gene-phenotype associations.
The basic concept behind our approach is to convert the numeric expression or ratio data into a genotype-like numeric format. With this data in hand, we can then use genome-wide association study (GWAS) tools such as GAPIT and GEMMA to conduct the association analysis between gene/exon expression or ratio and phenotype variation. This approach provides comprehensive results with GWAS.
Take the pod color TWAS with gene expression as an exmaple.
mkdir Exp
mv TWAS.MeasuredGeneExpression* Exp/
mv PodColor.ForAssociation.csv Exp/
cd Exp/
#rename the output file, so you can compare yours with it.
mv TWAS.MeasuredGeneExpression.PodColor.L2.CMLM.Results.csv TWAS.MeasuredGeneExpression.PodColor.L2.Publish.Results.csv
# If you already had R and the GAPIT package, then run below via terminal (Mac/Win) or any other IDE (like Rstudio)
Rscript TWAS.MeasuredGeneExpression.R # this will take couple of minutes
#Then you will have the output named with TWAS.MeasuredGeneExpression.PodColor.L2.CMLM.Results.csv, couple of commands to compare yours with mine
grep "Glyma.03G005700" | awk '{FS=","} print ($1,$4)}' *Results.csv # you should see identical results for the L2 gene, Glyma.03G005700, like below
TWAS.MeasuredGeneExpression.PodColor.L2.CMLM.Results.csv "Glyma.03G005700.Wm82.a2.v1" 1.15266722367601e-24
TWAS.MeasuredGeneExpression.PodColor.L2.Publish.Results.csv "Glyma.03G005700.Wm82.a2.v1" 1.15266722367726e-24
awk '{FS=","} {if( $4 <0.00001){print FILENAME,$0}}' *Results.csv
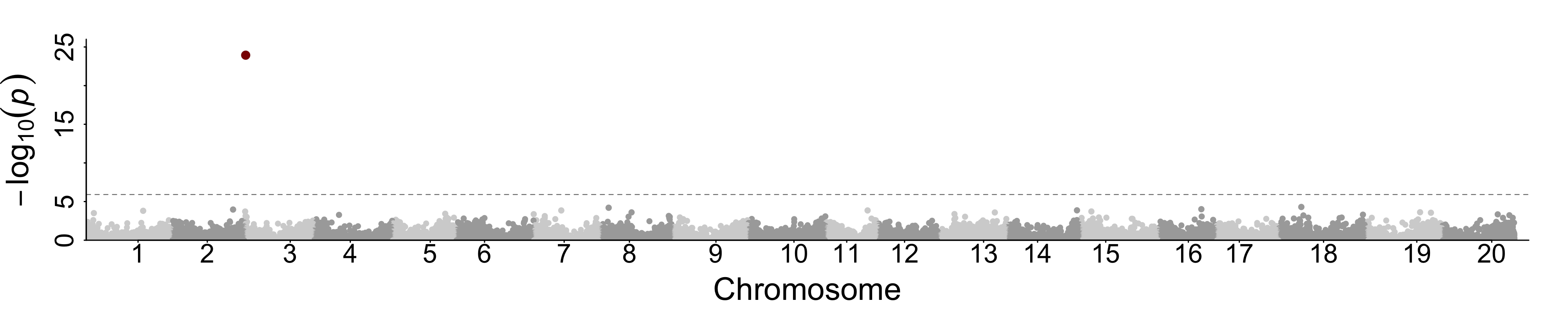
#see figure below
Note: The colnames of SNP is the default name from GAPIT, but here it's gene ID.
Below is the manhatton plot of Pod Color TWAS with measured gene expression.
I. Download the weight files of Soybean Seedling Shoot Cis-eQTL & Cis-exonQTL
mkdir FUSION
mv FUSION.* FUSION
cd FUSION/
tar -zxvf FUSION.LDREF.tar.gz
tar -zxvf eqtl.tar.gz
tar -zxvf LDREF.tar.gz
gunzip FUSION.PodColor.L2.GWAS.gemma.txt
II. Prepare your GWAS results with all input SNPs (NO Filtering). Four columns are required at least. Here, take output of GEMMA GWAS results as an example. And We provided pod color GWAS FUSION.PodColor.L2.GWAS.gemma.txt as an example
-
SNP– SNP identifier (rsID), therscolumn, please use Chr-Position as the SNP ID in order to be consistency with the reference panel LDREF. -
A1– first allele (effect allele),allele1of GEMMA assoc.txt -
A2– second allele (other allele),allele0of GEMMA assoc.txt -
Z– Z-scores,se/betaof GEMMA assoc.txt.
III. Intergrate the Cis-eQTL & Cis-exonQTL with your GWAS. A bash script to run FUSION by chromosome.
#!/usr/bin/bash
for chr in {1..20}
do
Rscript FUSION.assoc_test.R --sumstats FUSION.PodColor.L2.GWAS.gemma.txt --weights WEIGHTS.SeedlingShoot.pos --weights_dir WEIGHTS --ref_ld_chr LDREF/Gmax_eQTL_ --chr $chr --out L2.Fusion.${chr}.dat --max_impute 0.6
done
IV. for the output, you can use bonferronie cutoff for TWAS pvalue TWAS.P (≤0.05/eqtls) to define the associated genes. The files in out folder is the output that you supposed to get.
You can campare yours with ours, which was provided as FUSION.PodColor.Output.zip.
I. Download the summary-level data from the Soybean Seedling Shoot Cis-eQTL & Cis-exonQTL in binary format.
mkdir SMR
mv SMR.* SMR
cd SMR
tar -zxvf SMR.LDREF.tar.gz
tar -zxvf SMR.eqtl.tar.gz
gunzip GWAS/PodColor.L2.GWAS.gemma.ForFusion.txt.gz
II. Prepare your GWAS results with all your SNPs (NO Filtering). Eight columns are required. Here, take output of GEMMA GWAS results as an example
SNP– SNP identifier (rsID), therscolumn, please use Chr-Position as the SNP ID in order to be consistency with the reference panel LDREF.A1– alternative allele (effect allele),allele1of GEMMA assoc.txtA2– reference allele (other allele),allele0of GEMMA assoc.txtfreq– frequency of alternative allele (effect allele),afof GEMMA assoc.txtb–betaof GEMMA assoc.txt.se–seof GEMMA assoc.txt.p–p_waldof GEMMA assoc.txt.n– sample number for GWAS. you can setNAas not avalibale.
III. Intergrate the Cis-eQTL & Cis-exonQTL with your GWAS. Take the pod color GWAS as an exmaple.
smr --bfile LDREF/Gmax_eQTL --gwas-summary GWAS/PodColor.L2.assoc.gemma.ForSMR.txt --beqtl-summary eqtl/SeedlingShoot --out PodColor.L2.SMR
IV. The output file, you can use bonferronie cutoff for TWAS pvalue p_SMR (≤0.05/eqtls) and heidi test pvalue p_HEIDI ≥ 0.05 define the associated genes. Personal experience is that focus on p_SMR cutoff. p_HEIDI didn't help in our known genes.
You can campare your results with ours, which was provided as SMR.output.PodColor.L2.gz.
Welcome any question to make TWAS easily accessible for the plant community.
- My trait tissue is not related with the
seedling shoot, could I use this eQTL data? Answer: Yes. We did successfully get the known gene of trait from non-realted tissue, including disease resistance traits. It's certainly worth for a try. But you need to be careful with a possible higher false positive rate. - What do you mean Cis-eQTLs and Cis-exonQTLs? Answer: (1) eQTLs: genetic loci regulating gene expression (the trait); exonQTLs: genetic loci regulating exon reads proportion in certain gene. (2) We focused on Cis, which is likely the causal vairiations to decarese the false positives. (3) exonQTLs could capture alternative splicing and are more sensitive to structural virations (InDel, Gene fusion ...).
-
If you use the data of this respository, please cite our preprint: Li D, Wang Q, Tian Y et al. Transcriptome brings variations of gene expression, alternative splicing, and structural variations into gene-scale trait dissection in soybean. bioRxiv. 2023:2023-07.
-
The first soybean TWAS using meaured expression data, had conclusion that TWAS is roubust with source of expression data: Delin Li, Qiang Liu, Patrick S Schnable. TWAS results are complementary to and less affected by linkage disequilibrium than GWAS, Plant Physiology, Volume 186, Issue 4, August 2021, Pages 1800–1811, https://doi.org/10.1093/plphys/kiab161
-
Citation for software and method of SMR:
-
Citation for script and method of Fusion :
- SMR by Yang Lab
- Fusion by Gusev Lab
- GAPIT by Zhiwu Zhang Lab
- Thanks for support from National Natural Science Foundation of China (32201759) and National Key Research and Development Program of China (2021YFD1201601).