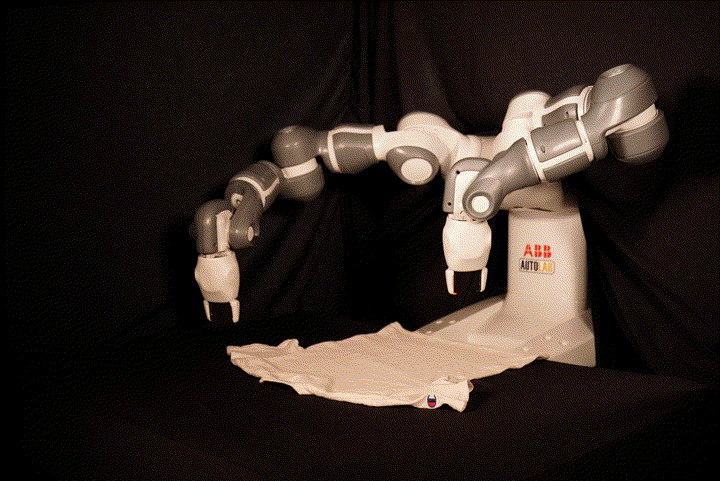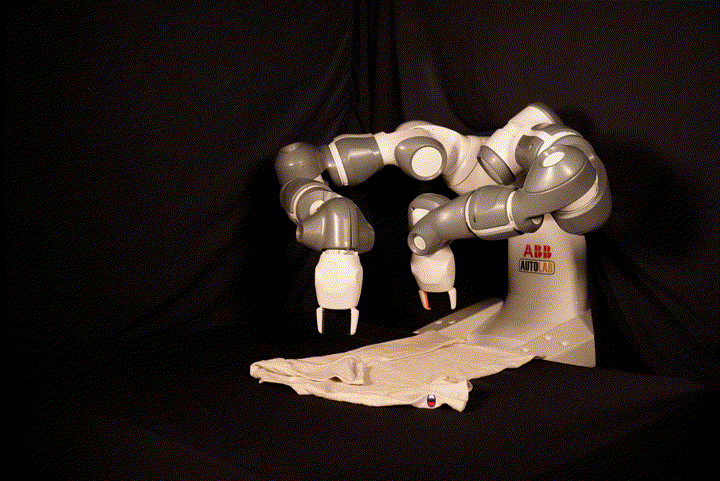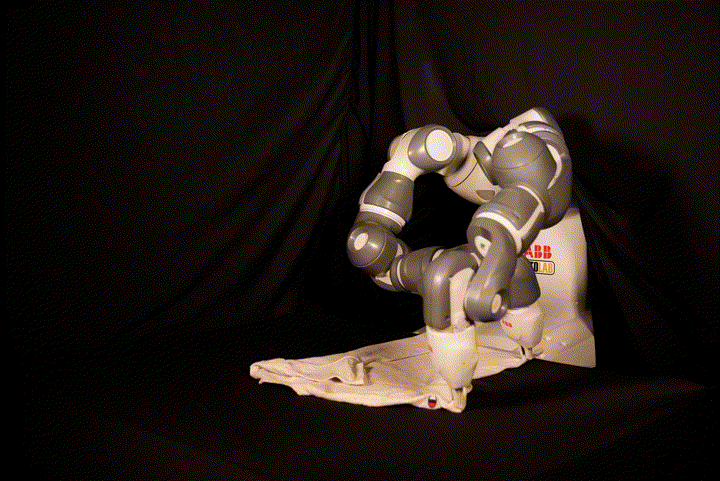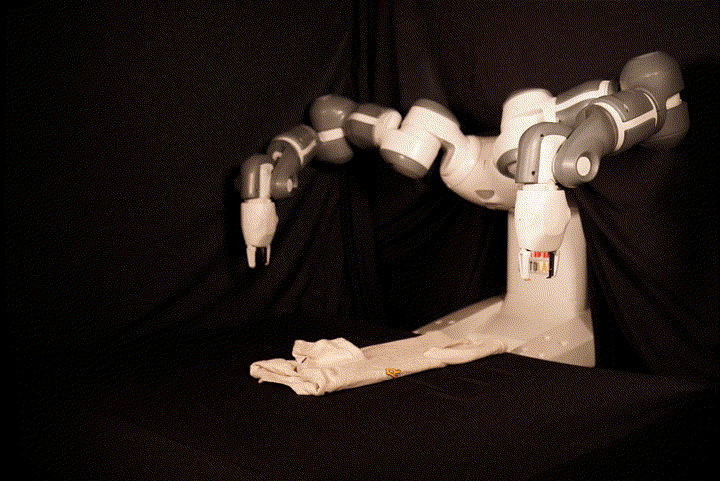
This repository contains the code for SpeedFolding: Learning Efficient Bimanual Folding of Garments, with the corresponding paper accepted at the 2022 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS 2022) in Kyoto. The code is not meant to be easily runnable, as it depends tightly on the hardware setup including the ABB Yumi robot and a Photoneo PhoXi camera. We recommend to see it as a reference implementation, in particular for our BiMama-Net architecture.
The SpeedFolding code depends on abb_librws, the SDK for Photoneo PhoXi camera, as well as on the relevant NVIDIA driver and CUDA installation for PyTorch. Further Python (3.6+) dependencies can then be installed via
pip install -r requirements.txt
pip install -e third_party/*
The third_party components, namely an OMPL planner and a PhoXi camera wrapper, are written in C++ and use pybind11 to expose Python funcionality. Before running the scripts, make sure to have both the learning and manipulation directory in your PYTHONPATH.
The overall structure is as follows:
/dataincludes the user-specified instructions for folding garments and calibration data./databasecontains the backend-server for saving data to our database. Have a look atlearning/database.pyfor information about reading the database./learningis about our introduced BiMama-Net architecture (and everything related to training and running inference with it). It includes scripts for drawing, augmentation, as well as model and reward definitions.inference.pyis the main class for calculating predictions and the next action./manipulationincludes everything related to the robot and running the overall high-level pipeline. Whileyumi.pycontrols the robot itself,experiment.pydefines the motions for the manipulation primitives. Theheuristicsdirectory includes all calculations of primitives that are not learned, in particular for instruction matching or folding primitives.
- First, start the database by running
uvicorn main:app --app-dir databasefrom the project root directory. When running the Speedfolding setup, it will upload the most recent images to the database as the current image for debugging purposes. - To check robot motions or run single manipulation primitives, use
manipulation/experiment.py. For example,python manipulation/experiment.py --do-the-flingwill fling the garment from pre-defined pick poses.python manipulation/experiment.py --instruction shirtwill fold an already smooth shirt according to the defined folding-lines instruction.python manipulation/experiment.py --do-2s-foldwill apply the 2 seconds folding heuristic. - To run the complete pipeline, use
mainpulation/run.py. Most of our experiments were run using thepython manipulation/run.py --horizon 10 --demo --foldarguments. To repeat the experiment even after a robot error, swaprun.pywithrun_forever.pykeeping the same arguments. The overall end-to-end pipeline depends on a number of hyperparameters that are explained furthermore.
| Group | Parameter | Commonly used value |
|---|---|---|
| Database | URL | http://127.0.0.1:8000 |
| Camera | PhoXi Serial number | 1703005 |
| Extrinsic calibration | data/calibrations/phoxi_to_world_bww.tf | |
| Motion | Speed (Full) | 0.6 m/s |
| Speed (Half) | 0.12 m/s | |
| Speed (Fling) | 1.0 m/s | |
| Speed (Stretch) | 0.06 m/s | |
| Force Threshold (Stretch) | 0.025 N | |
| Timeout | 7 s | |
| Grasping | Inwards distance | 0.018 m |
| Inwards angle | 0.25 rad | |
| Gripper force | 20 N | |
| Approach distance | 0.04 m | |
| Learning | Inference image size | 256 x 192 px |
| Depth image distances | 0.85 - 1.25 m | |
| Reachability masks | data/masks/{left, right}.npy | |
| Bimama-Net rotations | 20 | |
| Bimama-Net Embedding size | 8 | |
| Training epochs | 100 | |
| Learning rate | 4e-4 | |
| Learning rate exponential decay | 0.97 |
These are some of the parameters. In particular, the parametrization of the mainipulation primitives are too complex to state here. We refer to the manipulation/experiment.py directly.