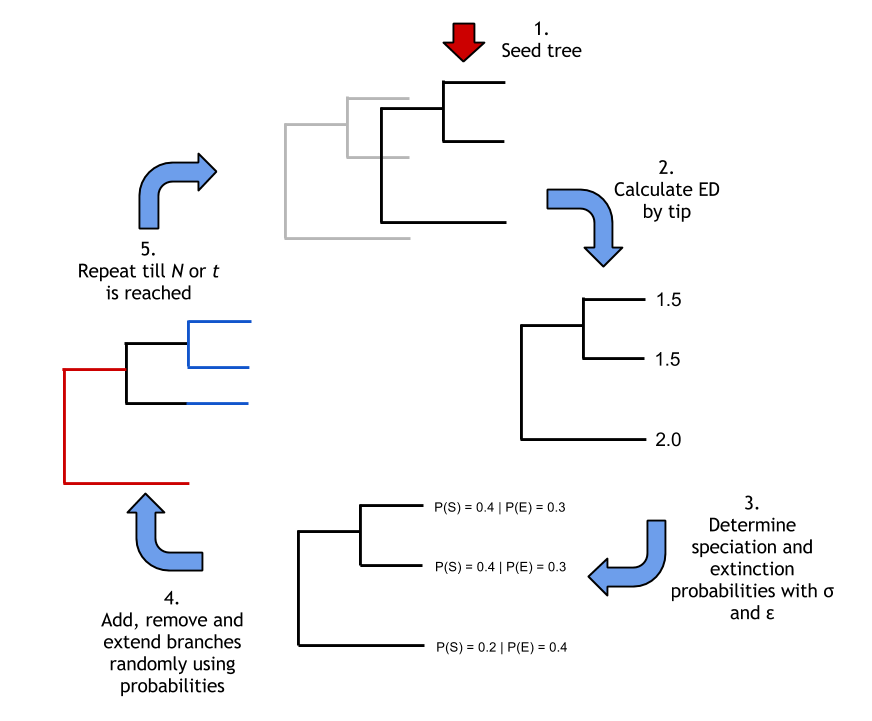
An R pipeline for exploring how an evolutionary distinctness bias in a tree-growth Markov model affects tree shape in order to test the reality of the living fossil, see the published article 'Evolutionarily distinct “living fossils” require both lower speciation and lower extinction rates'.
The pipeline uses a Markov-Model to grow trees with extinction and speciation rates determined by a tip's evolutionary distinctness.
Data and results files are not provided in this repository, only the code is. All results and starting datasets can be found in the Dryad repository. The repository holds a large (5GB) compressed file in five parts. These parts must all be downloaded, combined and then uncompressed to produce the folders.
# recombine and uncompress (UNIX)
cat xaa xab xac xad xae > data_results.tar.gz
tar zxvf data_results.tar.gz
System requirements
Run the install_deps.R script to install all dependent packages automatically.
- OS
- UNIX (but readily modifiable for Windows)
- R version
- 3+
- R packages (version used)
plyr(1.8.1)ape(3.2)geiger(2.0.3)apTreeshape(1.4.5)ggplot2(1.0.0)MoreTreeTools(0.0.1)outliers(0.14)doMC(1.3.3, not for Windows)foreach(1.4.2)test_that(0.9.1, optional)
- External
Please note, MoreTreeTools is in development and can only be installed via GitHub. It is best to install using the EDBMM branch:
library(devtools)
install_github('DomBennett/MoreTreeTools', ref='edbmm') # install edbmm branch
Directory structure
-- data/
---- raw_trees/
------ literature/
-------- [manually added]
------ treebase/
---- parsed_trees/
---- treestats/
-- parameters/
---- [all analysis parameters]
-- stages/
---- [all stage .R scripts]
-- tools/
---- [all tool .R scripts]
-- results/
----- [all folders named by analysis as specified in run.R]
-- other/
-- sanity_checks/
-- PATHd8
----- [excutable if parsing trees]
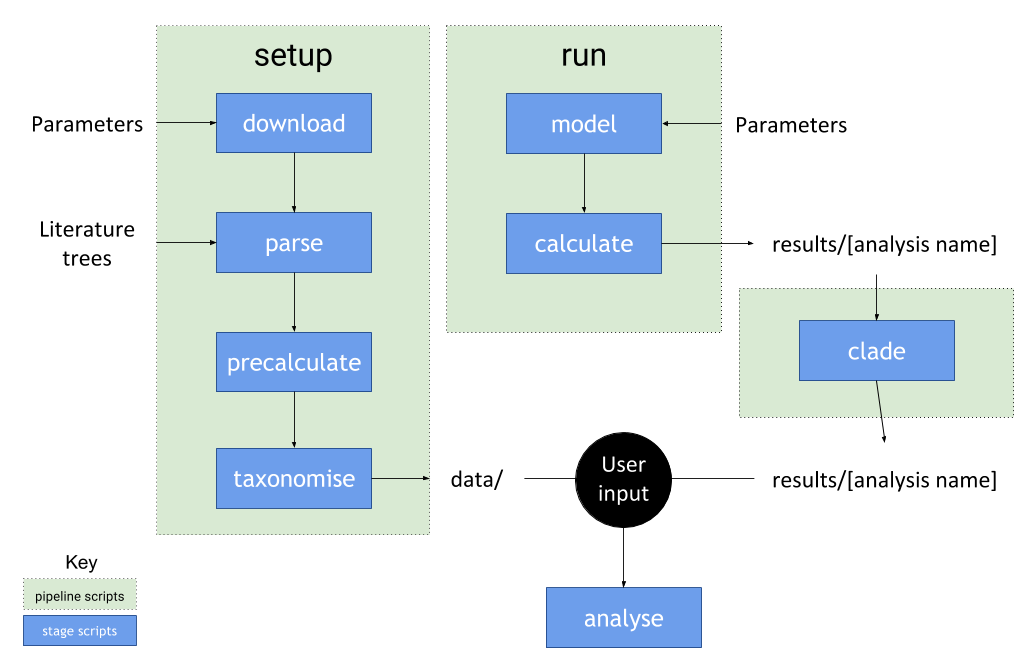
Pipeline
The pipeline works by calling the pipeline scripts setup.R and run.R. These scripts call
stage scripts which can be found in /stages.
The stages scripts depend on custom functions found in the /tools folder.
setup.R
This phase of the pipeline sources and calculates statistics from empirical trees:
- Download trees from TreeBase
- Parse trees (make ultramteric using different methods)
- Calculate tree shape statistics
- Determine the taxonomic identities of downloaded trees
run.R
This phase models trees for determining how model parameters affect tree shape:
- Read in parameters from
parameters/ - Model trees according to parameters in
run.R - Calculate tree shape statistics
Analysis
The final stage script is user interactive. All plots and analysis presented in the publication are generated with this script.
Clade analysis
In addition to setup and run, there are additional clade analysis stages: clade.R and analyse_clades.R.
Testing
Run test.R to make sure core functions are working.
Reference
Bennett, D.J., Sutton, M.D. & Turvey, S.T., 2016. Evolutionarily distinct “living fossils” require both lower speciation and lower extinction rates. Paleobiology, pp.1–15. Available at: http://www.journals.cambridge.org/abstract_S0094837316000361.
Author
Dom Bennett