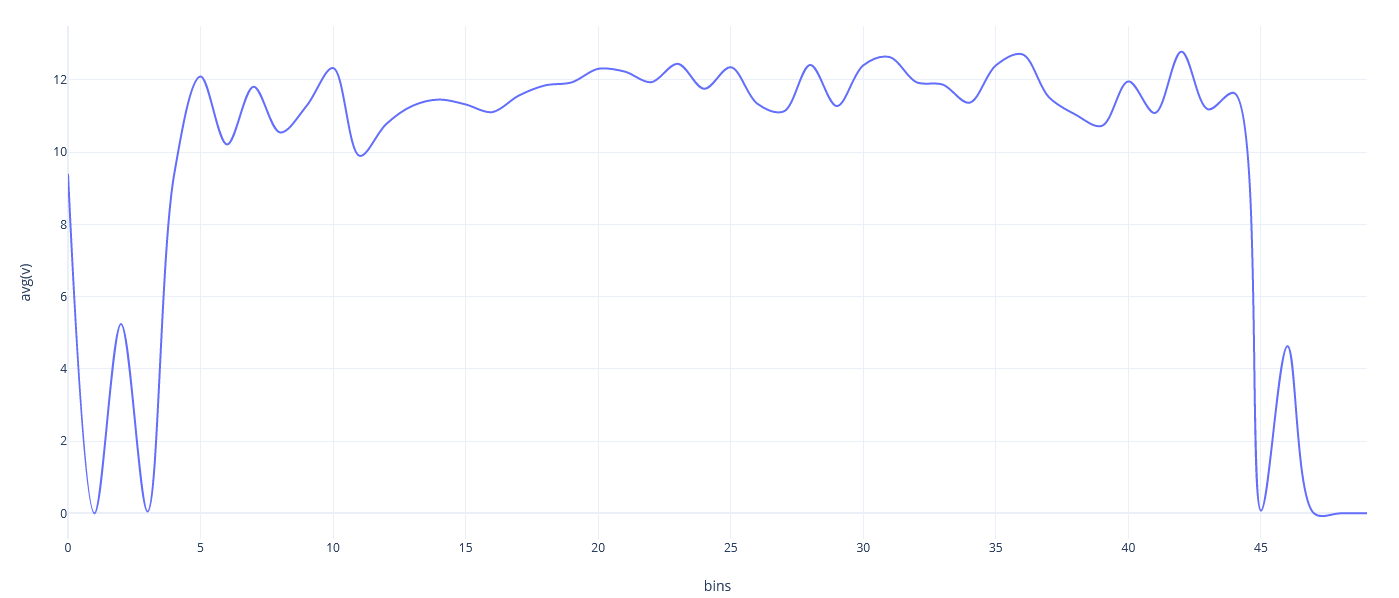
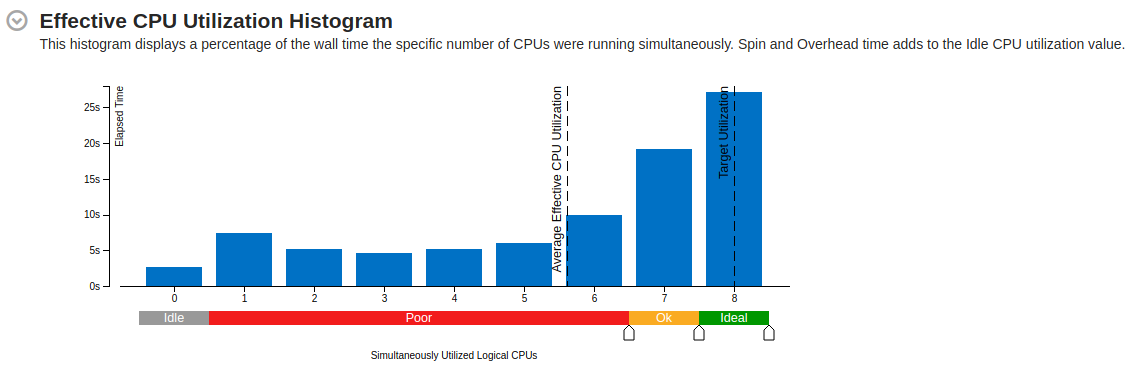
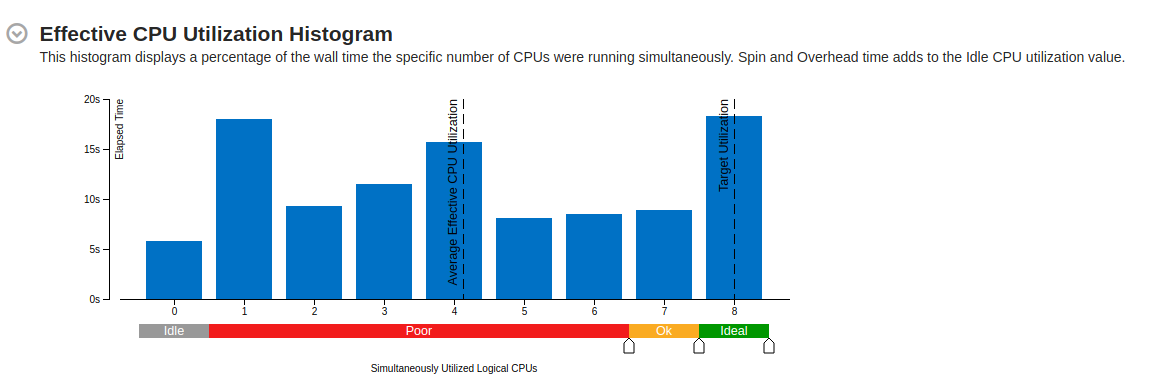
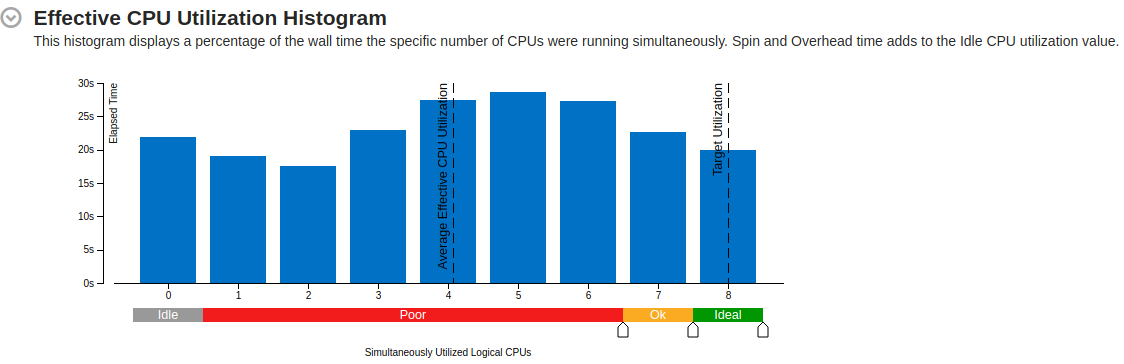
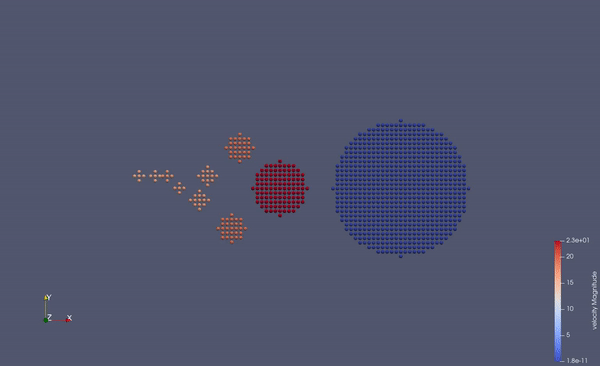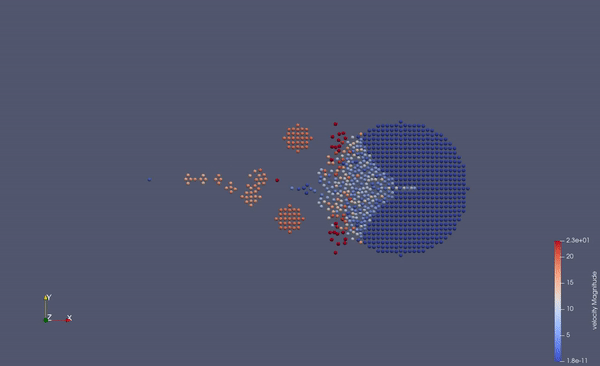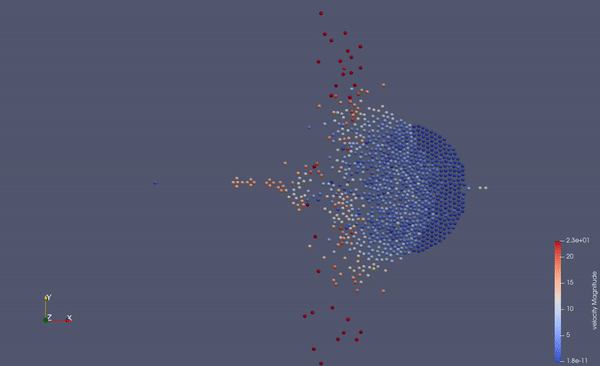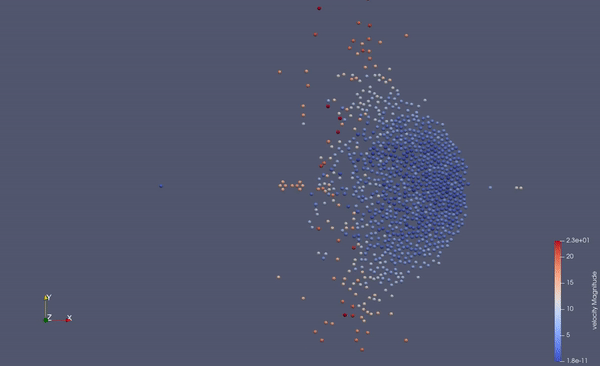
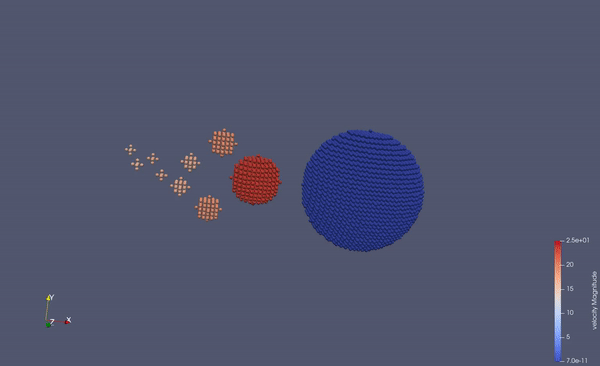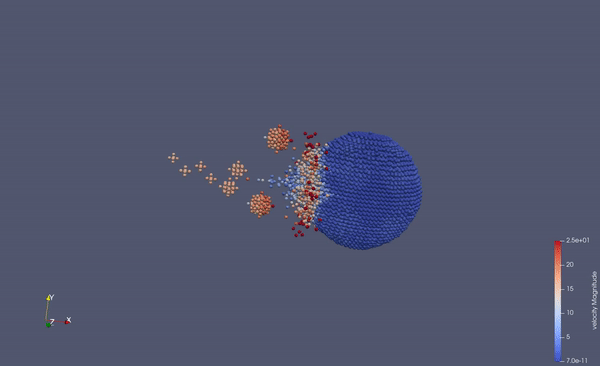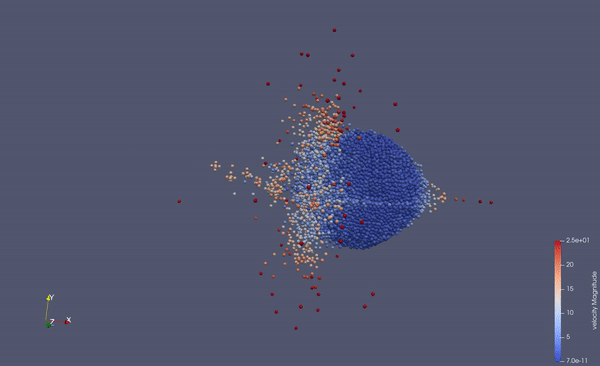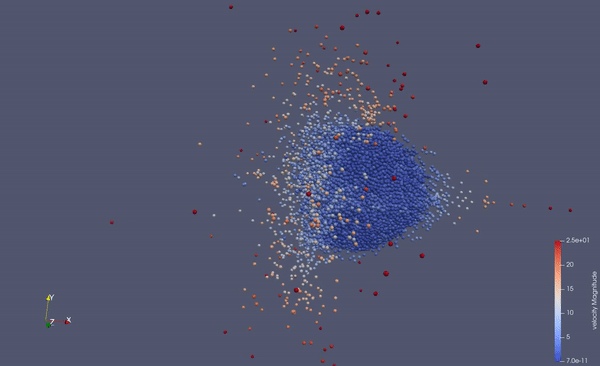
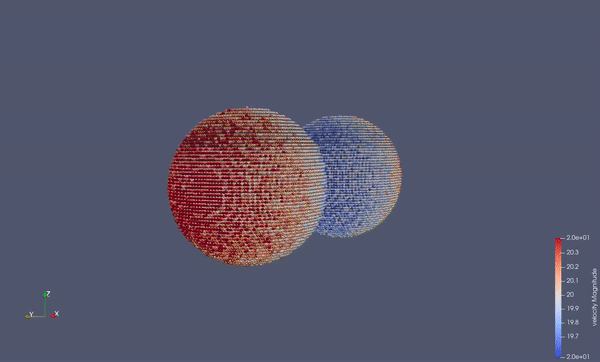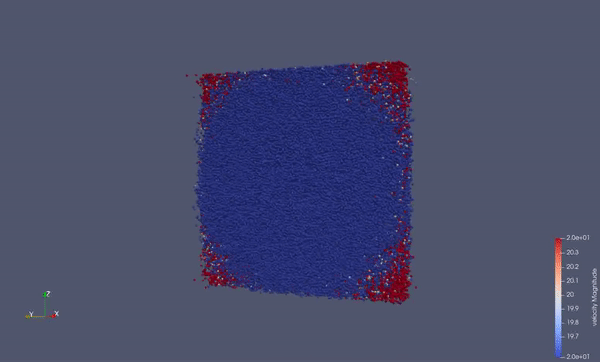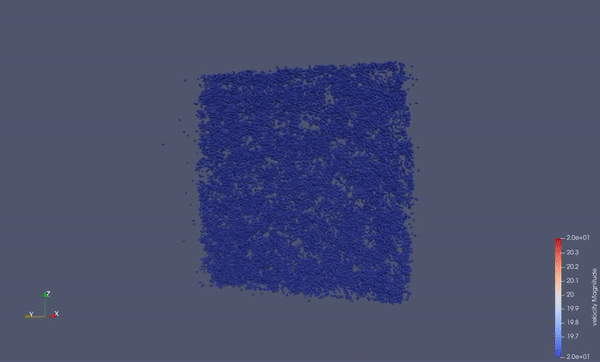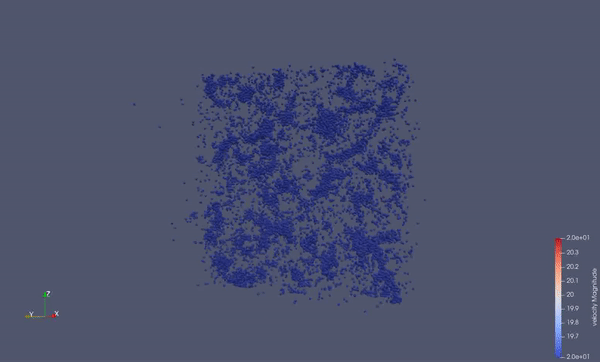
Part of a practical course on molecular dynamics at TU Munich (IN0012). Based on a template provided at TUM-I5/MolSim.
Application for molecular simulations.
Checkout the sources.
$ git clone git@github.com:Dominik-Weinzierl/MolSim.git
$ cd MolSimThe building process can take a while! Please be patient.
BUILD_DOCUMENTATION: Enables build of doxygen documentation (default: off)BUILD_TESTS: Enable build of tests (default: off)
-
Create
buildfolder and run cmake with make.$ make
*Existing
buildfolder will be deleted and created again. -
Switch into your build folder.
$ cd ./build -
Create the
MolSimtarget with the generated Makefile.$ make
- Create
buildfolder (in-source-buildsare disabled).$ mkdir ./build
- Switch into your
buildfolder.$ cd ./build - Run
cmakewith specified arguments.$ cmake .. -D BUILD_DOCUMENTATION=OFF -D BUILD_TESTS=OFF -D CMAKE_C_COMPILER=gcc -D CMAKE_CXX_COMPILER=g++
- Create the MolSim target with the generated Makefile.
$ make
To perform simulations in
2Dor3D, please add the corresponding flag (-2|-3) as an argument. For more details see the following examples.
Run ./MolSim without any arguments to list possible and required arguments.
$ ./MolSim
Usage: ./MolSim [-h | --help] | {-f | --filename} <filename> {-t | --t_end} <t_end> {-d | --delta_t} <delta_t> [-o | --output <output>] [-i | --iteration <iteration>] [-w | --writer {vtk | xyz}] [-p | --physics {gravitation | lennard}] [-b | --benchmark] [-2 | -3]
Options:
-h,--help Show this help message
-f,--filename Specify the input filename
-t,--t_end Specify the end time of this simulation
-d,--delta_t Specify the time steps per calculation
-o,--output Specify the output filename
-i,--iteration Specify the iteration
-w,--writer Specify the writer used for the output files
-p,--physics Specify the physics used for the simulation
-b,--benchmark Run simulation as benchmark
-2,-3 Specify the domain of the simulation (default: 3D)
Usage: ./MolSim {-x | --xml} {-f | --filename <filename>} [-b | --benchmark] [-2 | -3]
Options:
-f,--filename Specify the input filename as xml
-b,--benchmark Run simulation as benchmark
-2,-3 Specify the domain of the simulation (default: 3D)
- Run example simulation of
Task 3.$ ./MolSim --filename ../../input/ws_01/eingabe-sonne.txt --t_end 1000 --delta_t 0.014 --physics gravitation

- (optional) Run additional simulation of the solar system.
$ ./MolSim -f ../../input/ws_01/sun_system.txt -t 1000 -d 0.014 -p gravitation
- (optional) Run example simulation of
Task 3as benchmark.$ ./MolSim --filename ../../input/ws_01/eingabe-sonne.txt --t_end 1000 --delta_t 0.014 --physics gravitation --benchmark
- Run example simulation of
Task 3.$ ./MolSim -x -f ../../input/ws_02/input_task_3.xml -2

- (optional) Run example simulation of
Task 3as benchmark.$ ./MolSim -x -f ../../input/ws_02/input_task_3.xml -b -2
- (optional) Run example simulation of
Task 3as3Dsimulation.$ ./MolSim -x -f ../../input/ws_02/input_task_3.xml -3
- Input file used for simulation of
Task 3.<Simulation endTime="5" deltaT="0.0002" iteration="60" physics="lennard" writer="vtk" output="MD"> <Shapes> <Cuboid mass="1.0" distance="1.1225" meanValue="0.1" packed="true" depthOfPotentialWell="5" zeroCrossing="1"> <Position x="0.0" y="0.0" z="0.0"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="40" y="8" z="1"/> </Cuboid> <Cuboid mass="1.0" distance="1.1225" meanValue="0.1" packed="true" depthOfPotentialWell="5" zeroCrossing="1"> <Position x="15.0" y="15.0" z="0.0"/> <Velocity x="0.0" y="-10.0" z="0.0"/> <Dimension x="8" y="8" z="1"/> </Cuboid> </Shapes> </Simulation>
- Run example simulation of
Task 2.$ ./MolSim -x -f ../../input/ws_03/task_2_linked.xml -2
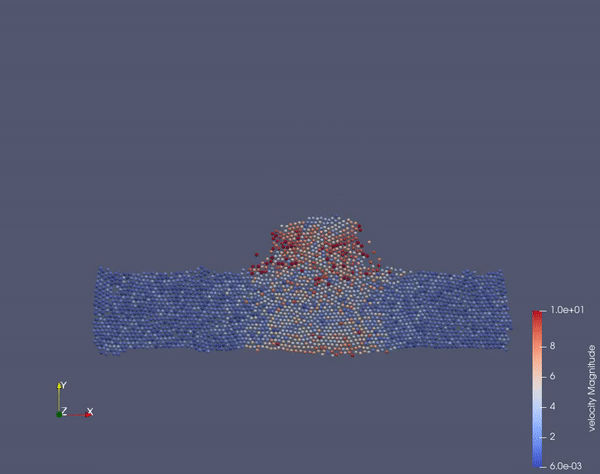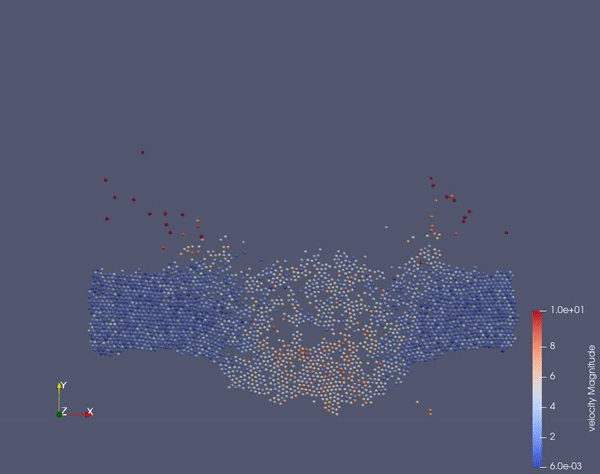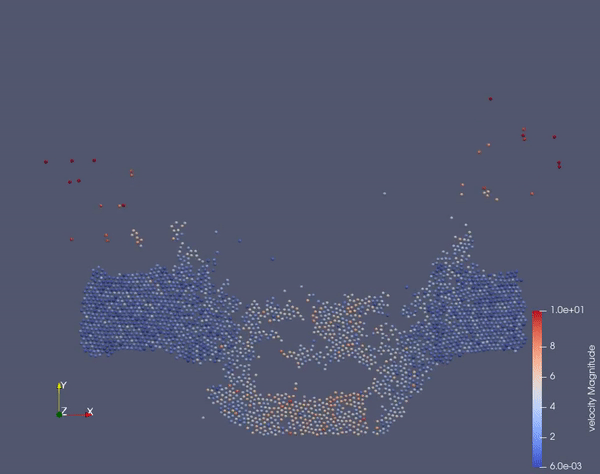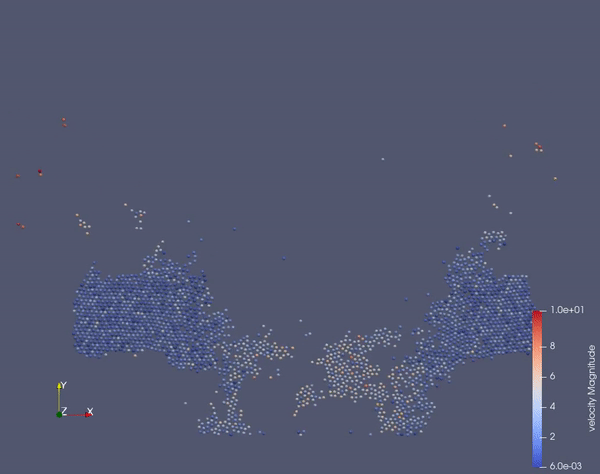
- (optional) Run example simulation of
Task 2as direct sum simulation.$ ./MolSim -x -f ../../input/ws_03/task_2_direct.xml -2
- (optional) Run example simulation of
Task 2as benchmark.$ ./MolSim -x -f ../../input/ws_03/task_2_linked.xml -b -2
- (optional) Run example simulation of
Task 2as3Dsimulation.$ ./MolSim -x -f ../../input/ws_03/task_2_linked.xml -3
- Input file used for simulation of
Task 2.<Simulation endTime="20" deltaT="0.0005" iteration="60" physics="lennard" writer="vtk" output="MD"> <Strategy> <LinkedCell cutoffRadius="3"> <Domain x="180" y="90" z="9"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary="outflow"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.1225" meanValue="0.1" packed="true" depthOfPotentialWell="5" zeroCrossing="1"> <Position x="20.0" y="20.0" z="1.0"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="100" y="20" z="1"/> </Cuboid> <Cuboid mass="1.0" distance="1.1225" meanValue="0.1" packed="true" depthOfPotentialWell="5" zeroCrossing="1"> <Position x="70" y="60" z="1.0"/> <Velocity x="0.0" y="-10.0" z="0.0"/> <Dimension x="20" y="20" z="1.0"/> </Cuboid> </Shapes> </Simulation>
- Run example simulation of
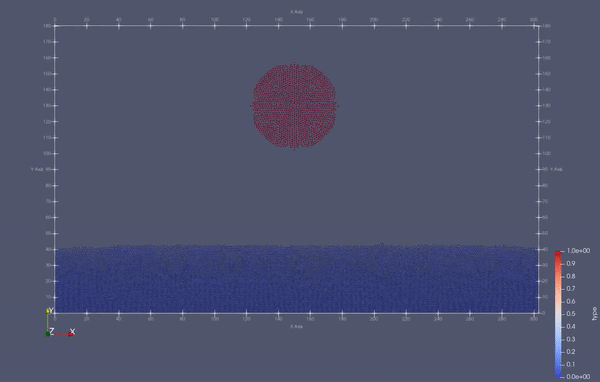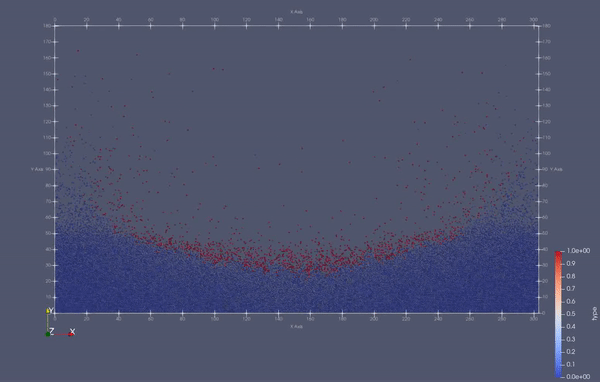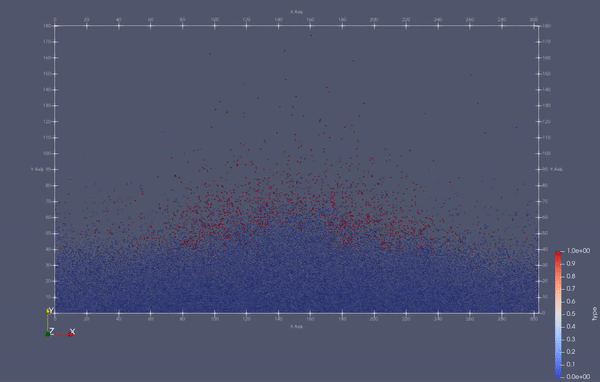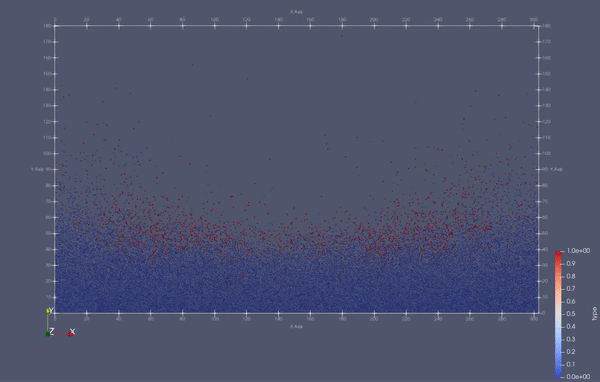
Task 4.$ ./MolSim -x -f ../../input/ws_03/water_drop.xml -2
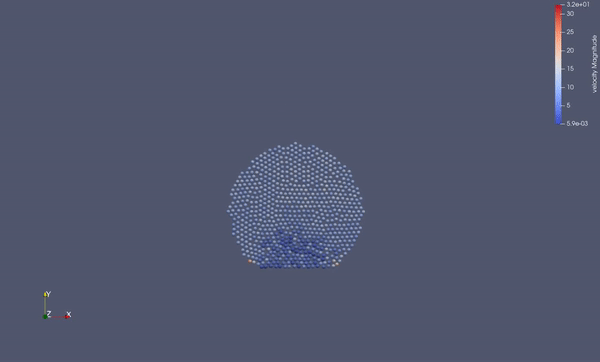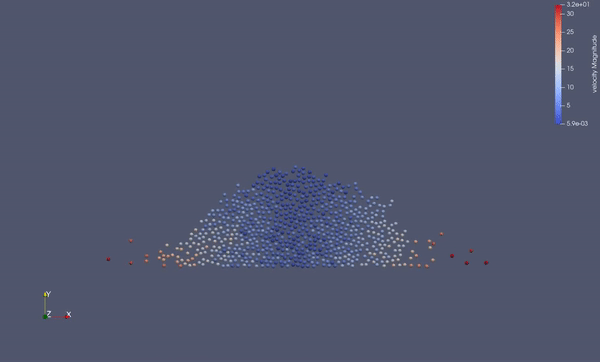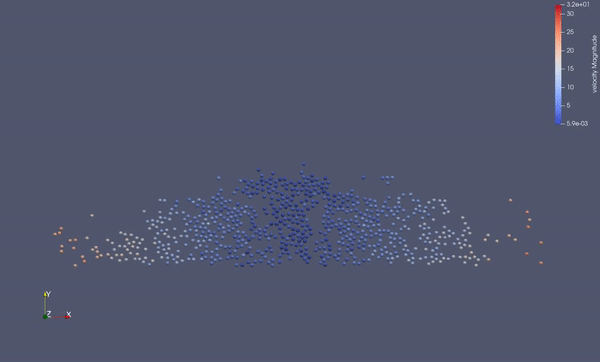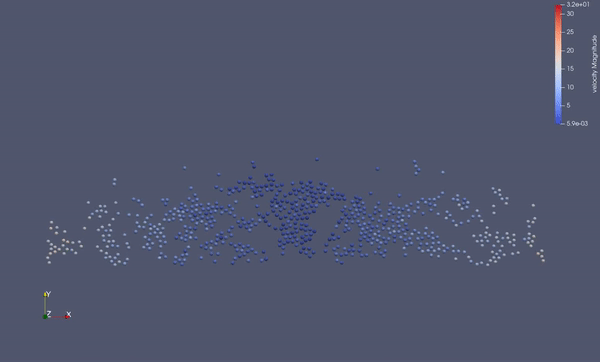
- (optional) Run example simulation of
Task 4as benchmark.$ ./MolSim -x -f ../../input/ws_03/water_drop.xml -b -2
- (optional) Run example simulation of
Task 4as3Dsimulation.$ ./MolSim -x -f ../../input/ws_03/water_drop.xml -3
- Input file used for simulation of
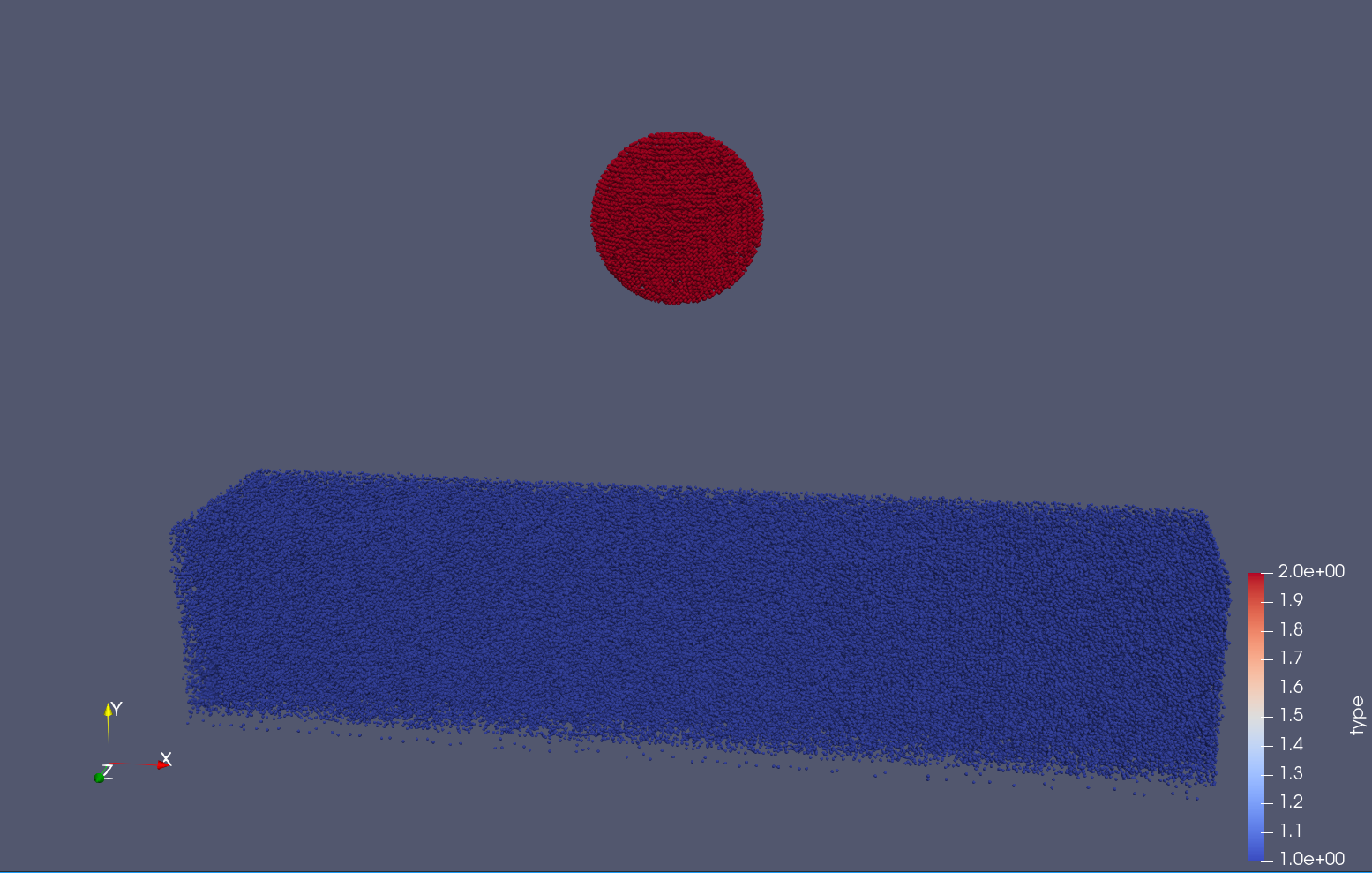
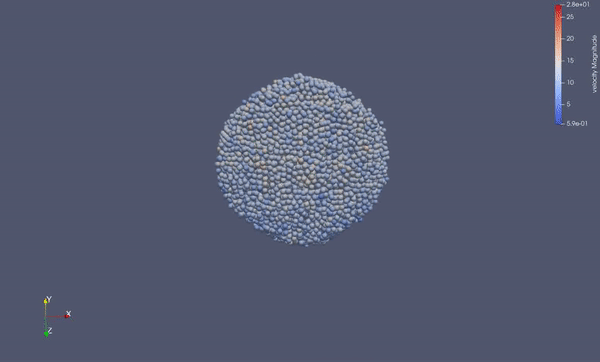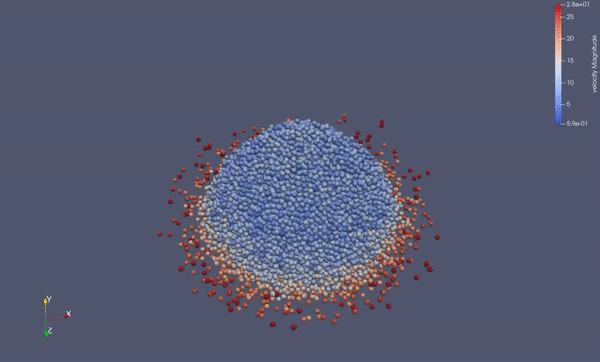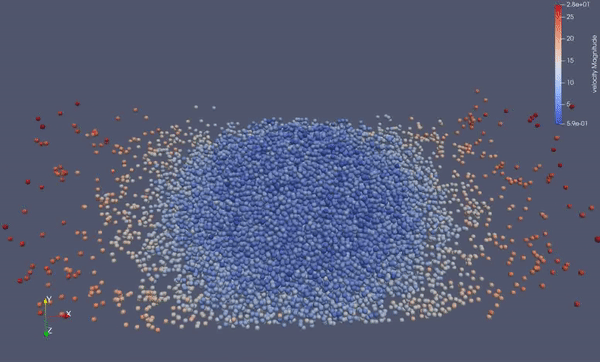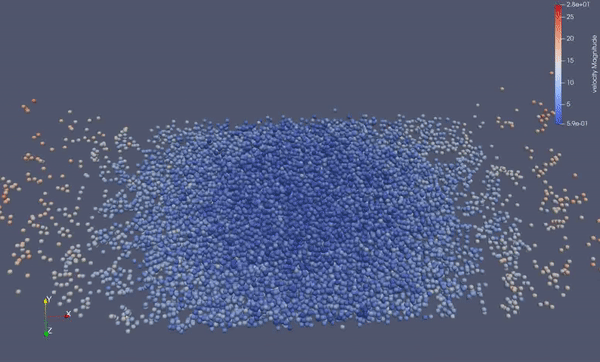
Task 4.<Simulation endTime="10" deltaT="0.00005" iteration="120" physics="lennard" writer="vtk" output="MD"> <Strategy> <LinkedCell cutoffRadius="3"> <Domain x="120" y="51" z="51"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-bottom="reflecting"/> </LinkedCell> </Strategy> <Shapes> <Sphere mass="1.0" distance="1.1225" meanValue="0.1" radius="15" packed="true" depthOfPotentialWell="5" zeroCrossing="1"> <Center x="60.0" y="25.0" z="25.0"/> <Velocity x="0.0" y="-10.0" z="0.0"/> </Sphere> </Shapes> </Simulation>
-
Run example simulation of
Task 2(small).$ ./MolSim -x -f ../../input/ws_04/task_2_small.xml -2
-
(optional) Run example simulation of
Task 2as benchmark.$ ./MolSim -x -f ../../input/ws_04/task_2_small.xml -b -2
-
(optional) Run example simulation of
Task 2as3Dsimulation.$ ./MolSim -x -f ../../input/ws_04/task_2_small.xml -3
-
Input file used for simulation of
Task 2.<Simulation endTime="25" deltaT="0.0005" iteration="120" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x ="0" y="-12.44" z="0"/> <Strategy> <LinkedCell cutoffRadius="2.5" parallel="lock-optimized"> <Domain x="63" y="36" z="63"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="reflecting" boundary-bottom="reflecting" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1" type="0"> <Position x="0.6" y="2.0" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="14" z="50"/> </Cuboid> <Cuboid mass="2.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="0.9412" type="1"> <Position x="0.6" y="19.0" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="14" z="50"/> </Cuboid> </Shapes> <Thermostat initialT="40" numberT="1000"/> </Simulation>
-
Run example simulation of
Task 2(big).$ ./MolSim -x -f ../../input/ws_04/task_2_big.xml -2
-
(optional) Run example simulation of
Task 2as benchmark.$ ./MolSim -x -f ../../input/ws_04/task_2_big.xml -b -2
-
(optional) Run example simulation of
Task 2as3Dsimulation.$ ./MolSim -x -f ../../input/ws_04/task_2_big.xml -3
-
Input file used for simulation of
Task 2.<Simulation endTime="10" deltaT="0.0005" iteration="120" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x="0" y="-12.44" z="0"/> <Strategy> <LinkedCell cutoffRadius="3" parallel="lock-optimized"> <Domain x="300" y="54" z="300"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="reflecting" boundary-bottom="reflecting" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.2" type="0"> <Position x="0.6" y="2.0" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="250" y="20" z="250"/> </Cuboid> <Cuboid mass="2.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.1" type="1"> <Position x="0.6" y="27.0" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="250" y="20" z="250"/> </Cuboid> </Shapes> <Thermostat initialT="40" numberT="1000"/> </Simulation>
-
Run example simulation of
Task 3.$ ./MolSim -x -f ../../input/ws_04/task_3_fluid.xml -2
-
(optional) Run example simulation of
Task 3as benchmark.$ ./MolSim -x -f ../../input/ws_04/task_3_fluid.xml -b -2
-
(optional) Run example simulation of
Task 3as3Dsimulation.$ ./MolSim -x -f ../../input/ws_04/task_3_fluid.xml -3
-
Input file used for simulation of
Task 3.<Simulation endTime="15" deltaT="0.0005" iteration="120" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x ="0" y="-12.44" z="0"/> <Strategy> <LinkedCell cutoffRadius="3"> <Domain x="303" y="180" z="72"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="reflecting" boundary-bottom="reflecting" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.2" type="1"> <Position x="1.5" y="2.0" z="1.5"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="250" y="50" z="50"/> </Cuboid> </Shapes> <Thermostat initialT="0.5" numberT="1000"/> </Simulation>
-
Run example simulation of
Task 3.$ ./MolSim -x -f ../../input/ws_04/task_3_checkpointing.xml -2
-
(optional) Run example simulation of
Task 3as benchmark.$ ./MolSim -x -f ../../input/ws_04/task_3_checkpointing.xml -b -2
-
(optional) Run example simulation of
Task 3as3Dsimulation.$ ./MolSim -x -f ../../input/ws_04/task_3_checkpointing.xml -3
Note that you may need to recreate the
task_3_fluid_calm.vtufile. -
Input file used for simulation of
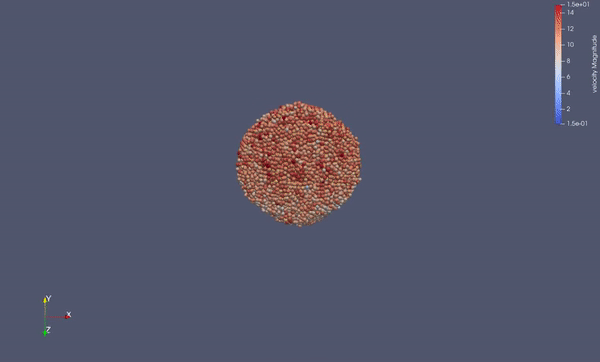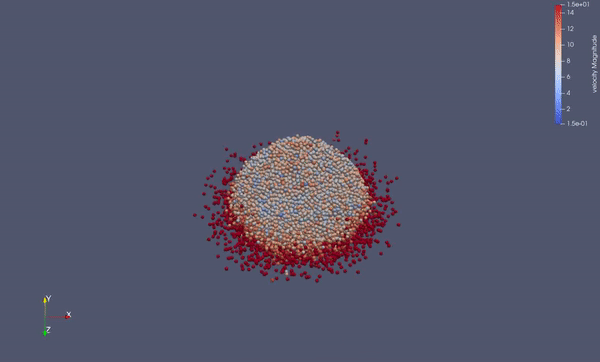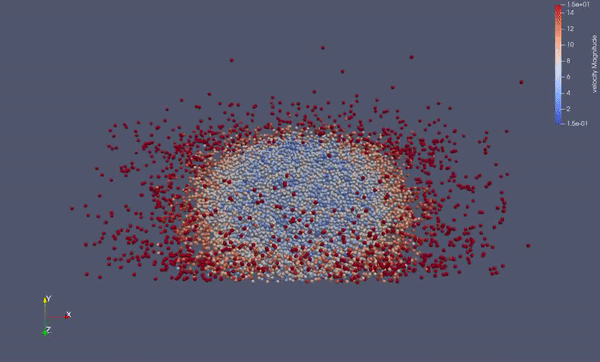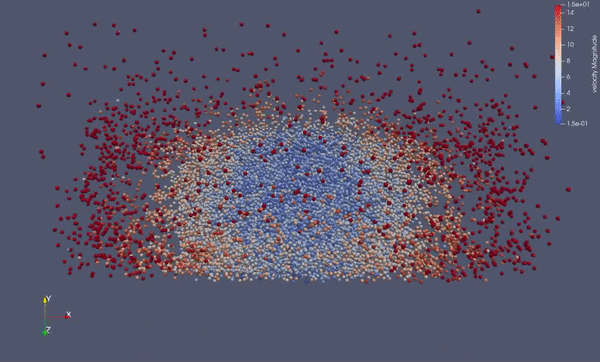
Task 3.<Simulation endTime="40" deltaT="0.0005" iteration="120" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x ="0" y="-12.44" z="0"/> <Source path="../../input/ws_04/task_3_fluid_calm.vtu"/> <Strategy> <LinkedCell cutoffRadius="3"> <Domain x="303" y="180" z="72"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="reflecting" boundary-bottom="reflecting" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Sphere mass="1.0" distance="1.2" meanValue="0.0" radius="20" packed="true" depthOfPotentialWell="1.0" zeroCrossing="1.2" type="1"> <Center x="150.0" y="150.0" z="35"/> <Velocity x="0.0" y="0.0" z="0.0"/> </Sphere> </Shapes> </Simulation>
- Run example simulation of
Task 1.$ ./MolSim -x -f ../../input/ws_05/task_1.xml -3

- (optional) Run example simulation of
Task 1as benchmark.$ ./MolSim -x -f ../../input/ws_05/task_1.xml -b -3
- Input file used for simulation of
Task 1.<Simulation endTime="500" deltaT="0.01" iteration="60" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x="0.0" y="0.0" z="-0.001"/> <Strategy> <LinkedCell cutoffRadius="4.0"> <Domain x="148" y="148" z="148"/> <CellSize x="4" y="4" z="4"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="periodic" boundary-bottom="periodic" boundary-front="reflecting" boundary-back="reflecting"/> </LinkedCell> </Strategy> <Shapes> <Membrane mass="1.0" distance="2.2" meanValue="0" packed="true" depthOfPotentialWell="1.0" zeroCrossing="1.0" stiffness="300" averageBondLength="2.2" type="0"> <Position x="15.0" y="15.0" z="1.5"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="50" z="1"/> <Forces start="0" end="150"> <Strength x="0.0" y="0.0" z="0.8"/> <Index x="17" y="24" z="0"/> <Index x="17" y="25" z="0"/> <Index x="18" y="24" z="0"/> <Index x="18" y="25" z="0"/> </Forces> </Membrane> </Shapes> </Simulation>
-
Run example simulation of
Task 3.$ ./MolSim -x -f ../../input/ws_05/task_3.xml -3
-
(optional) Run example simulation of
Task 3as benchmark.$ ./MolSim -x -f ../../input/ws_05/task_3.xml -b -3
-
(optional) Run example simulation of
Task 3as2Dsimulation.$ ./MolSim -x -f ../../input/ws_05/task_3.xml -2
-
Input file used for simulation of
Task 3.<Simulation endTime="100" deltaT="0.0005" iteration="120" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x="0.0" y="-12.44" z="0.0"/> <Strategy> <LinkedCell cutoffRadius="3.6" parallel="lock-cell"> <Domain x="60" y="60" z="60"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="reflecting" boundary-bottom="reflecting" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.2" type="0"> <Position x="0.6" y="0.6" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="20" z="50"/> </Cuboid> <Cuboid mass="2.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.1" type="1"> <Position x="0.6" y="24.6" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="20" z="50"/> </Cuboid> </Shapes> <Thermostat initialT="40" numberT="1000"/> </Simulation>
-
Run example simulation of
Task 4.$ ./MolSim -x -f ../../input/ws_05/task_4.xml -3
-
(optional) Run example simulation of
Task 4as benchmark.$ ./MolSim -x -f ../../input/ws_05/task_4.xml -b -3
-
(optional) Run example simulation of
Task 4as2Dsimulation.$ ./MolSim -x -f ../../input/ws_05/task_4.xml -2
-
Input file used for simulation of
Task 4.<Simulation endTime="500" deltaT="0.0005" iteration="1000" physics="lennard" writer="vtk" output="NanoFlow"> <AdditionalGravitation x="0.0" y="-0.8" z="0.0"/> <Strategy> <LinkedCell cutoffRadius="2.75" parallel="lock-cell"> <Domain x="30" y="30" z="12"/> <CellSize x="3" y="3" z="3"/> <Boundary boundary-left="outflow" boundary-right="outflow" boundary-top="periodic" boundary-bottom="periodic" boundary-front="periodic" boundary-back="periodic"/> </LinkedCell> </Strategy> <Shapes> <Cuboid mass="1.0" distance="1.0" meanValue="0" packed="true" depthOfPotentialWell="2" zeroCrossing="1.1" type="1" fixed="true"> <Position x="1" y="0.5" z="0.5"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="2" y="30" z="12"/> </Cuboid> <Cuboid mass="1.0" distance="1.0" meanValue="0" packed="true" depthOfPotentialWell="2" zeroCrossing="1.1" type="1" fixed="true"> <Position x="27.2" y="0.5" z="0.5"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="2" y="30" z="12"/> </Cuboid> <Cuboid mass="1.0" distance="1.2" meanValue="0" packed="true" depthOfPotentialWell="1" zeroCrossing="1.0" type="2" fixed="false"> <Position x="3.2" y="0.6" z="0.6"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="20" y="25" z="10"/> </Cuboid> </Shapes> <Thermostat initialT="40" numberT="10" flow="true"/> <ProfileWriter numOfBins="10" numOfIterations="1000" velocity="true" density="false"/> </Simulation>
Four different parallelization strategies:
With this strategy, the calculation is not applied directly to the particles of the other cells and the result is first stored in a buffer. In this way, the calculation can be parallelized and updating of the values is processed sequentially without locks.
#pragma omp declare reduction (merge : std::vector<std::pair<Particle<dim> *, Vector<dim>>> : omp_out.insert(omp_out.end(), omp_in.begin(), omp_in.end()))Usage:
<Strategy>
<LinkedCell cutoffRadius="4.0" parallel="buffer">
[...]
</LinkedCell>
</Strategy>With this strategy there is a lock for each cell, and with each write access the cell is then locked accordingly.
#pragma omp parallel for shared(cellContainer) default(none) schedule(static, 4)
for (size_t c = 0; c < cellContainer.getBoundaryAndInnerCells().size(); ++c) {
Cell<dim> *cell = cellContainer.getBoundaryAndInnerCells()[c];
std::vector<Cell<dim> *> &neighbours = cell->getNeighbours();
std::vector<Particle<dim> *> &cellParticles = cell->getParticles();
if (!cellParticles.empty()) {
// calc between particles in cells and relevant neighbours
cell->setLock();
for (auto n = neighbours.begin(); n != neighbours.end(); ++n) {
(*n)->setLock();
[...]
(*n)->unsetLock();
}
cell->unsetLock();
cell->setLock();
LinkedCell<LennardJones, dim>::calcInTheCell(cellParticles, cellContainer);
cell->unsetLock();
std::vector<std::tuple<Cell<dim> *, Vector<dim>>> &periodicNeighbours = cell->getPeriodicNeighbours();
// Iterate over all periodic neighbours
for (std::tuple<Cell<dim> *, Vector<dim>> &t: periodicNeighbours) {
// Get the periodic cell which influences the current cell
Cell<dim> *periodicCell = std::get<0>(t);
if (!periodicCell->getParticles().empty()) {
periodicCell->setLock();
for (auto j = periodicCell->getParticles().begin(); j != periodicCell->getParticles().end(); ++j) {
// Update the current position of the Particle(s)
[...]
cell->setLock();
[...]
cell->unsetLock();
(*j)->setX(oldPos);
}
periodicCell->unsetLock();
}
}
}
}
[...]
}Usage:
<Strategy>
<LinkedCell cutoffRadius="4.0" parallel="lock-cell">
[...]
</LinkedCell>
</Strategy>With this strategy, no locks are used and the mesh is divided in such a way that no race conditions occur. Periodic neighbours are proceeded sequentially.
template<>
LinkedCellParallelLockFree<LennardJones, 2>::LinkedCellParallelLockFree(double cutoffRadius, Vector<2> cellSize,
ParticleContainer<2> &particleContainer) {
auto rangeX = std::ceil(cutoffRadius / cellSize[0]);
auto rangeY = std::ceil(cutoffRadius / cellSize[1]);
auto maxX = static_cast<size_t>(rangeX + rangeX);
auto maxY = static_cast<size_t>(rangeY + 1 + rangeY);
auto &cellContainer = dynamic_cast<LinkedCellContainer<2> &>(particleContainer);
auto cellsPerColumn = static_cast<unsigned long>(cellContainer.cellsPerColumn());
for (size_t x = 0; x < maxX; ++x) {
for (size_t y = 0; y < maxY; ++y) {
std::vector<Cell<2> *> cellVector;
size_t row = x;
size_t counter = row * cellsPerColumn + y;
while (counter < cellContainer.getBoundaryAndInnerCells().size()) {
cellVector.push_back(cellContainer.getBoundaryAndInnerCells()[counter]);
if (counter + maxY >= ((row + 1) * cellsPerColumn)) {
row += maxX;
counter = row * cellsPerColumn + y;
} else {
counter += maxY;
}
}
cells.push_back(cellVector);
}
}
}for (auto &cellVector: cells) {
#pragma omp parallel for shared(cellVector, cellContainer) default(none) schedule(static, 8)
for (size_t c = 0; c < cellVector.size(); ++c) {
Cell<dim> *cell = cellVector[c];
std::vector<Cell<dim> *> &neighbours = cell->getNeighbours();
std::vector<Particle<dim> *> &cellParticles = cell->getParticles();
if (!cellParticles.empty()) {
LinkedCell<LennardJones, dim>::calcBetweenNeighboursAndCell(neighbours, cellParticles, cellContainer);
LinkedCell<LennardJones, dim>::calcInTheCell(cellParticles, cellContainer);
}
}
}
// Periodic neighbours sequentially
for (size_t c = 0; c < cellContainer.getBoundaryAndInnerCells().size(); ++c) {
Cell<dim>* cell = cellContainer.getBoundaryAndInnerCells()[c];
std::vector<Particle<dim> *> &cellParticles = cell->getParticles();
LinkedCell<LennardJones, dim>::calcPeriodic(cellParticles, cellContainer, *cell);
}
[...]
}Usage:
<Strategy>
<LinkedCell cutoffRadius="4.0" parallel="lock-free">
[...]
</LinkedCell>
</Strategy>With this strategy, the mesh is divided as lock free, but in addition the periodic neighbours are processed in parallel with locks.
for (auto &cellVector: LinkedCellParallelLockFree<LennardJones, dim>::cells) {
#pragma omp parallel for shared(cellVector, cellContainer) default(none) schedule(static, 4)
for (size_t c = 0; c < cellVector.size(); ++c) {
[...]
if (!cellParticles.empty()) {
[....]
std::vector<std::tuple<Cell<dim> *, Vector<dim>>> &periodicNeighbours = cell->getPeriodicNeighbours();
// Iterate over all periodic neighbours
for (std::tuple<Cell<dim> *, Vector<dim>> &t: periodicNeighbours) {
// Get the periodic cell which influences the current cell
Cell<dim> *periodicCell = std::get<0>(t);
if (!periodicCell->getParticles().empty()) {
periodicCell->setLock();
[...]
periodicCell->unsetLock();
}
}
}
}
}Usage:
<Strategy>
<LinkedCell cutoffRadius="4.0" parallel="lock-optimized">
[...]
</LinkedCell>
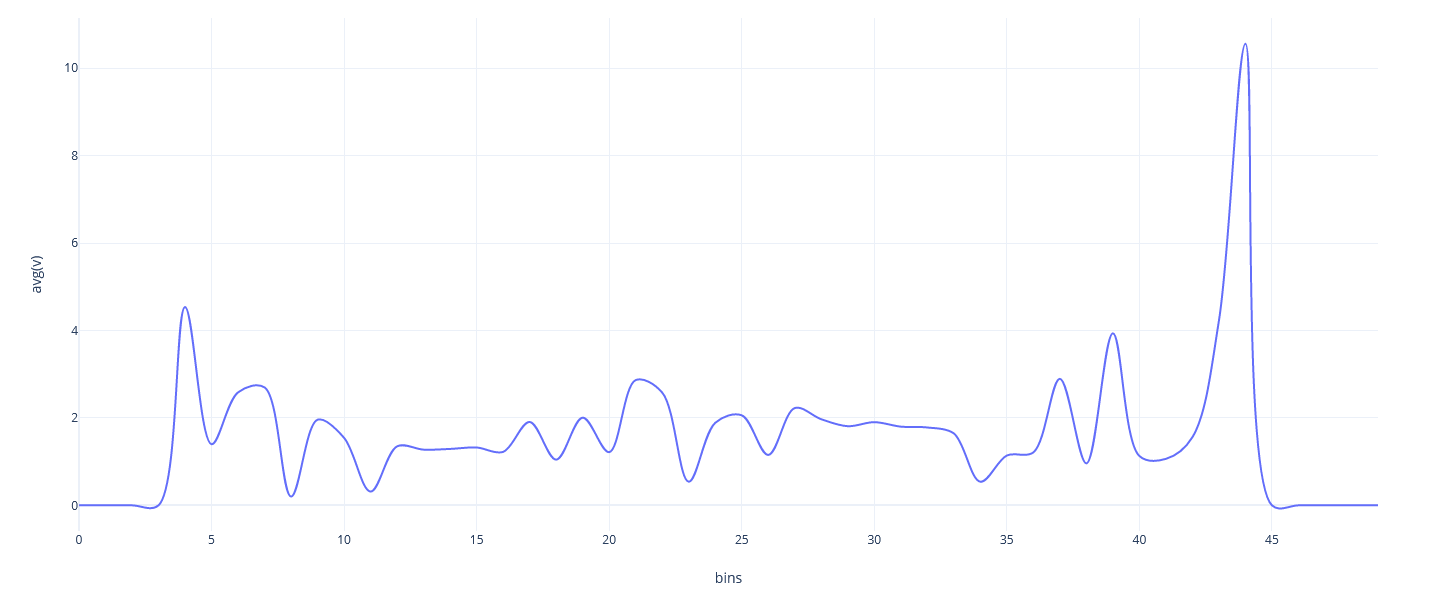
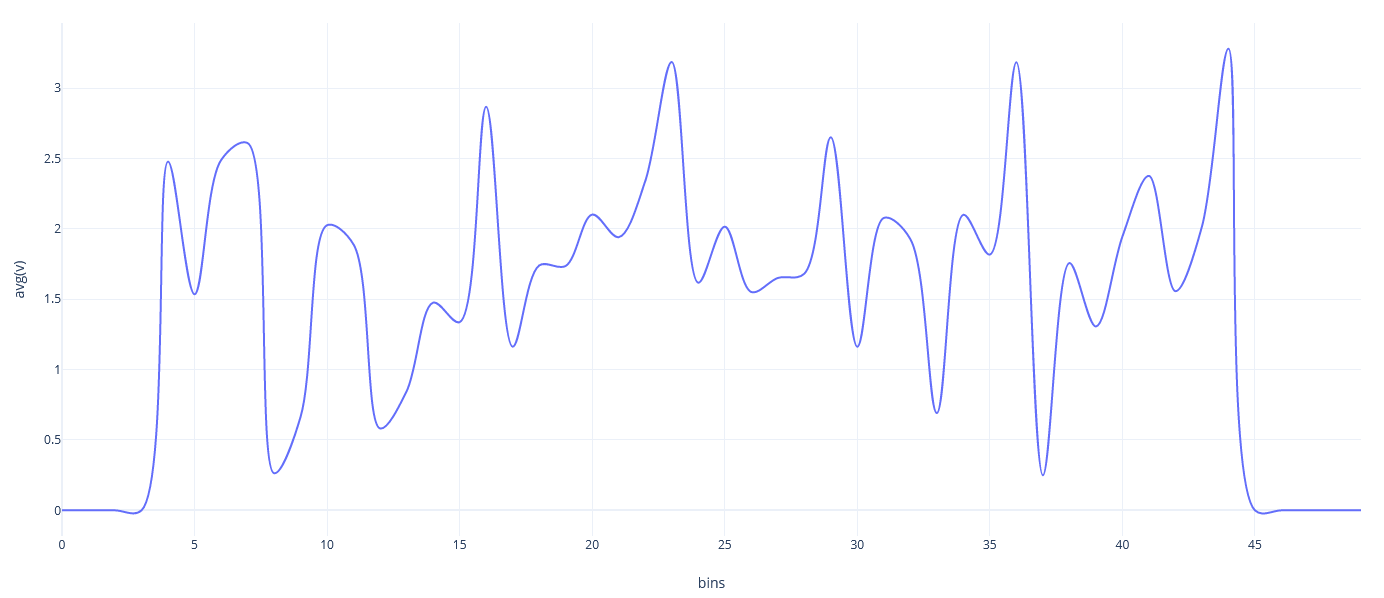
</Strategy>Example simulation of Worksheet 2 (Task 2 big) with 1, 2, 4, 8, 14, 16 and 28 Threads.
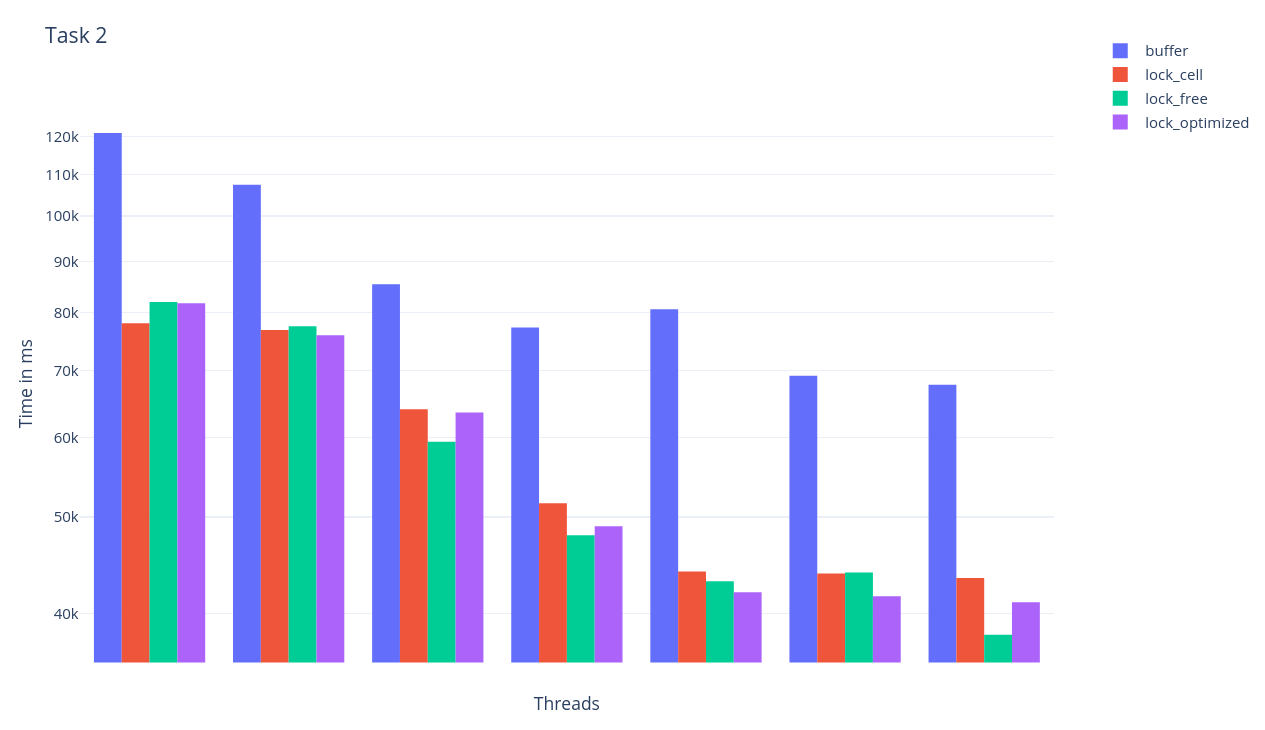
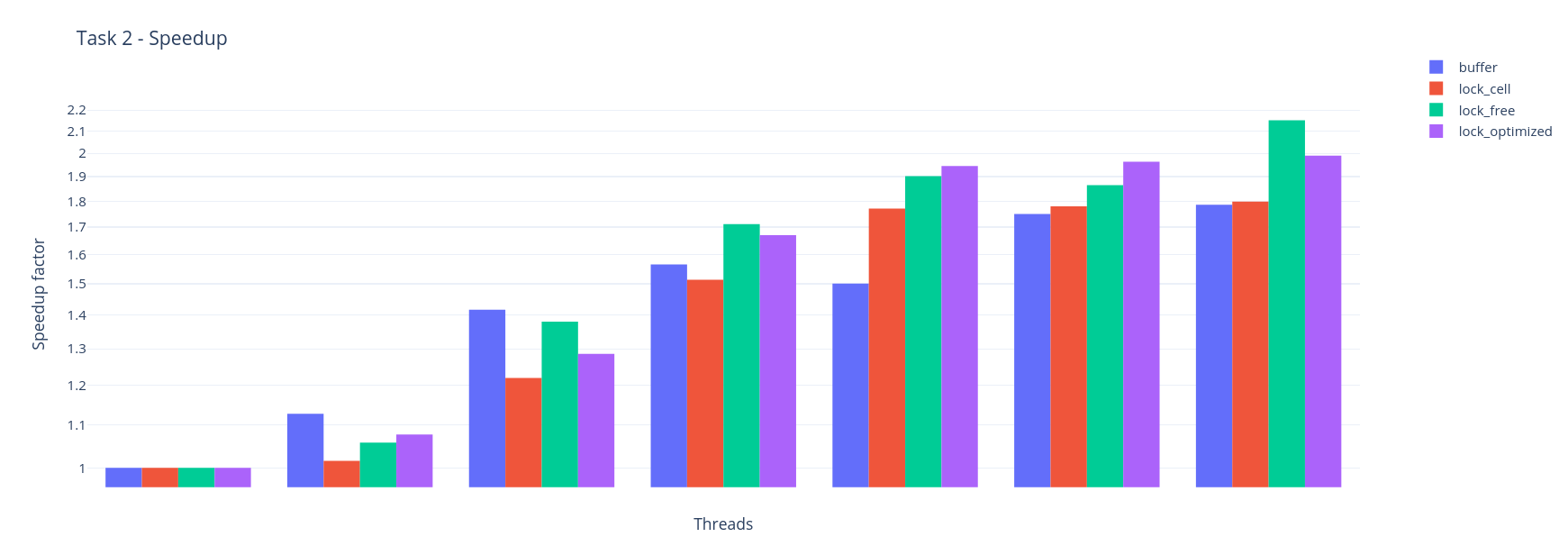
Disable all spdlog outputs to get best results. Therefore add -D WITH_SPD_LOG_OFF=OFF to your cmake command. (
Default: disabled)
Additional cmake options:
WITH_SPD_LOG_OFF"Disable logger" ONWITH_SPD_LOG_TRACE"Log level: Trace" OFFWITH_SPD_LOG_DEBUG"Log level: Debug" OFFWITH_SPD_LOG_INFO"Log level: Info" OFFWITH_SPD_LOG_WARN"Log level: Warn" OFFWITH_SPD_LOG_ERROR"Log level: Error" OFFWITH_SPD_LOG_CRITICAL"Log level: Critical" OFF
- Run
cmakeinbuildfolder with specified arguments.$ cmake .. -D BUILD_DOCUMENTATION=OFF -D BUILD_TESTS=ON -D CMAKE_C_COMPILER=gcc -D CMAKE_CXX_COMPILER=g++
- Run
makeinsourcefolder with specified arguments.$ make build_with_test
- Run
makeinbuildfolder to build target.$ make
- Run tests in
buildfolder to verify correctness.$ ctest [...] 100% tests passed, 0 tests failed out of 193 Total Test time (real) = 34.03 sec
-
XSD - Definition of xml file structure
<?xml version="1.0"?> <xsd:schema xmlns:xsd="http://www.w3.org/2001/XMLSchema"> <!-- Cuboids --> <xsd:complexType name="cuboid_t"> <xsd:sequence> <xsd:element name="Position" type="vector_t"/> <xsd:element name="Velocity" type="vector_t"/> <xsd:element name="Dimension" type="vector_i"/> <xsd:element name="Forces" type="force_t" minOccurs="0" maxOccurs="unbounded"/> </xsd:sequence> <xsd:attribute name="fixed" type="xsd:boolean"/> <xsd:attribute name="distance" type="xsd:double" use="required"/> <xsd:attribute name="mass" type="xsd:double" use="required"/> <xsd:attribute name="meanValue" type="xsd:double" use="required"/> <xsd:attribute name="packed" type="xsd:boolean" use="required"/> <xsd:attribute name="depthOfPotentialWell" type="xsd:double" use="required"/> <xsd:attribute name="zeroCrossing" type="xsd:double" use="required"/> <xsd:attribute name="type" type="xsd:nonNegativeInteger"/> </xsd:complexType> <!-- Sphere --> <xsd:complexType name="sphere_t"> <xsd:sequence> <xsd:element name="Center" type="vector_t"/> <xsd:element name="Velocity" type="vector_t"/> <xsd:element name="Forces" type="force_t" minOccurs="0" maxOccurs="unbounded"/> </xsd:sequence> <xsd:attribute name="fixed" type="xsd:boolean"/> <xsd:attribute name="radius" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="distance" type="xsd:double" use="required"/> <xsd:attribute name="mass" type="xsd:double" use="required"/> <xsd:attribute name="meanValue" type="xsd:double" use="required"/> <xsd:attribute name="packed" type="xsd:boolean" use="required"/> <xsd:attribute name="depthOfPotentialWell" type="xsd:double" use="required"/> <xsd:attribute name="zeroCrossing" type="xsd:double" use="required"/> <xsd:attribute name="type" type="xsd:nonNegativeInteger"/> </xsd:complexType> <!-- Membrane --> <xsd:complexType name="membrane_t"> <xsd:sequence> <xsd:element name="Position" type="vector_t"/> <xsd:element name="Velocity" type="vector_t"/> <xsd:element name="Dimension" type="vector_i"/> <xsd:element name="Forces" type="force_t" minOccurs="0" maxOccurs="unbounded"/> </xsd:sequence> <xsd:attribute name="fixed" type="xsd:boolean"/> <xsd:attribute name="distance" type="xsd:double" use="required"/> <xsd:attribute name="mass" type="xsd:double" use="required"/> <xsd:attribute name="meanValue" type="xsd:double" use="required"/> <xsd:attribute name="packed" type="xsd:boolean" use="required"/> <xsd:attribute name="depthOfPotentialWell" type="xsd:double" use="required"/> <xsd:attribute name="zeroCrossing" type="xsd:double" use="required"/> <xsd:attribute name="stiffness" type="xsd:double" use="required"/> <xsd:attribute name="averageBondLength" type="xsd:double" use="required"/> <xsd:attribute name="type" type="xsd:nonNegativeInteger"/> <xsd:attribute name="fixedOutline" type="xsd:boolean"/> </xsd:complexType> <!-- Double vector --> <xsd:complexType name="vector_t"> <xsd:attribute name="x" type="xsd:double" use="required"/> <xsd:attribute name="y" type="xsd:double" use="required"/> <xsd:attribute name="z" type="xsd:double" use="required"/> </xsd:complexType> <!-- Integer vector --> <xsd:complexType name="vector_i"> <xsd:attribute name="x" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="y" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="z" type="xsd:nonNegativeInteger" use="required"/> </xsd:complexType> <!-- Force vector --> <xsd:complexType name="force_t"> <xsd:sequence> <xsd:element name="Strength" type="vector_t"/> <xsd:element name="Index" type="vector_i" minOccurs="0" maxOccurs="unbounded"/> </xsd:sequence> <xsd:attribute name="start" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="end" type="xsd:nonNegativeInteger" use="required"/> </xsd:complexType> <!-- Profile writer --> <xsd:complexType name="profilewriter_t"> <xsd:attribute name="numOfBins" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="numOfIterations" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="velocity" type="xsd:boolean" use="required"/> <xsd:attribute name="density" type="xsd:boolean" use="required"/> </xsd:complexType> <!-- Input --> <xsd:complexType name="input_t"> <xsd:attribute name="path" type="xsd:string" use="required"/> </xsd:complexType> <!-- Boundary options --> <xsd:complexType name="boundary_t"> <xsd:attribute name="boundary" type="xsd:string"/> <xsd:attribute name="boundary-right" type="xsd:string"/> <xsd:attribute name="boundary-left" type="xsd:string"/> <xsd:attribute name="boundary-top" type="xsd:string"/> <xsd:attribute name="boundary-bottom" type="xsd:string"/> <xsd:attribute name="boundary-back" type="xsd:string"/> <xsd:attribute name="boundary-front" type="xsd:string"/> </xsd:complexType> <!-- List of Shapes (Cuboids/Spheres) --> <xsd:complexType name="shape_t"> <xsd:sequence> <xsd:element name="Cuboid" type="cuboid_t" minOccurs="0" maxOccurs="unbounded"/> <xsd:element name="Sphere" type="sphere_t" minOccurs="0" maxOccurs="unbounded"/> <xsd:element name="Membrane" type="membrane_t" minOccurs="0" maxOccurs="unbounded"/> </xsd:sequence> </xsd:complexType> <!-- Direct sum simulation --> <xsd:complexType name="directSum_t"/> <!-- Linked cell simulation --> <xsd:complexType name="linkedCell_t"> <xsd:sequence> <!-- Boundary: specify at which boundary of the domain which type of condition is applied --> <xsd:element name="Boundary" type="boundary_t"/> <!-- Domain: size of the domain --> <xsd:element name="Domain" type="vector_t"/> <!-- CellSize: size of the cells --> <xsd:element name="CellSize" type="vector_t"/> </xsd:sequence> <!-- cutoffRadius: used for linked cell optimizations --> <xsd:attribute name="cutoffRadius" type="xsd:double" use="required"/> <xsd:attribute name="parallel" type="xsd:string"/> </xsd:complexType> <!-- Strategy selection --> <xsd:complexType name="strategy_t"> <xsd:choice> <xsd:element name="LinkedCell" type="linkedCell_t"/> <xsd:element name="DirectSum" type="directSum_t"/> </xsd:choice> </xsd:complexType> <!-- Thermostat --> <xsd:complexType name="thermostat_t"> <!-- cutoffRadius: used for linked cell optimizations --> <xsd:attribute name="initialT" type="xsd:double" use="required"/> <xsd:attribute name="targetT" type="xsd:double"/> <xsd:attribute name="numberT" type="xsd:nonNegativeInteger" use="required"/> <xsd:attribute name="deltaT" type="xsd:nonNegativeInteger"/> <xsd:attribute name="flow" type="xsd:boolean"/> </xsd:complexType> <!-- Simulation --> <xsd:complexType name="simulation_t"> <xsd:sequence> <xsd:element name="Shapes" type="shape_t" minOccurs="0" maxOccurs="unbounded"/> <xsd:element name="Source" type="input_t" minOccurs="0" maxOccurs="unbounded"/> <xsd:element name="Strategy" type="strategy_t" minOccurs="0"/> <xsd:element name="Thermostat" type="thermostat_t" minOccurs="0"/> <xsd:element name="AdditionalGravitation" type="vector_t" minOccurs="0"/> <xsd:element name="ProfileWriter" type="profilewriter_t" minOccurs="0"/> </xsd:sequence> <xsd:attribute name="endTime" type="xsd:double" use="required"/> <xsd:attribute name="deltaT" type="xsd:double" use="required"/> <xsd:attribute name="output" type="xsd:string" use="required"/> <xsd:attribute name="iteration" type="xsd:nonNegativeInteger" use="required"/> <!-- physics: gravitation | lennard --> <xsd:attribute name="physics" type="xsd:string" use="required"/> <!-- writer: vtk | xyz --> <xsd:attribute name="writer" type="xsd:string" use="required"/> </xsd:complexType> <xsd:element name="Simulation" type="simulation_t"/> </xsd:schema>
-
XML - Example input file
<Simulation endTime="500" deltaT="0.01" iteration="60" physics="lennard" writer="vtk" output="MD"> <AdditionalGravitation x="0.0" y="0.0" z="-0.001"/> <Strategy> <LinkedCell cutoffRadius="4.0"> <Domain x="148" y="148" z="148"/> <CellSize x="4" y="4" z="4"/> <Boundary boundary-left="periodic" boundary-right="periodic" boundary-top="periodic" boundary-bottom="periodic" boundary-front="reflecting" boundary-back="reflecting"/> </LinkedCell> </Strategy> <Shapes> <Membrane mass="1.0" distance="2.2" meanValue="0" packed="true" depthOfPotentialWell="1.0" zeroCrossing="1.0" stiffness="300" averageBondLength="2.2" type="0"> <Position x="15.0" y="15.0" z="1.5"/> <Velocity x="0.0" y="0.0" z="0.0"/> <Dimension x="50" y="50" z="1"/> <Forces start="0" end="150"> <Strength x="0.0" y="0.0" z="0.8"/> <Index x="17" y="24" z="0"/> <Index x="17" y="25" z="0"/> <Index x="18" y="24" z="0"/> <Index x="18" y="25" z="0"/> </Forces> </Membrane> </Shapes> </Simulation>
You need to perform the following commands in the top level project folder.
- Remove the
buildfolder.$ make clean
- Remove and create the
buildfolder.$ make create_folder
You need to perform the following commands in the build folder.
- Make the
doxygendocumentation.$ make doc_doxygen
- Remove the
doxygendocumentation.$ make clean_doxygen
- Remove all target relevant files (e.g. Target, ...).
$ make clean
Our project is developed by Dominik, Janin and Nils as part of Group A.
MolSim is released under the MIT license.
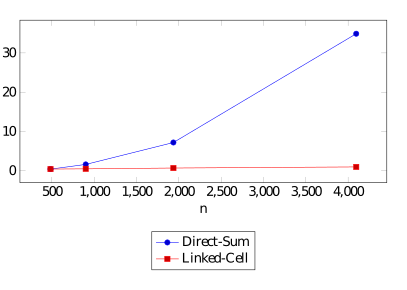
This simulation currently offers two strategy to perform ths results: The naive "Direct Sums" implementation and a smarter "Linked Cell" algorithm.
We ran some benchmarks and came to the following runtimes in ms/iteration for the two approaches depending on the number of particle in a 2D-square.