- Introduction
- Release note
- Requirements
- Installation
- Configuration
- Usage
- Output files
- Modules
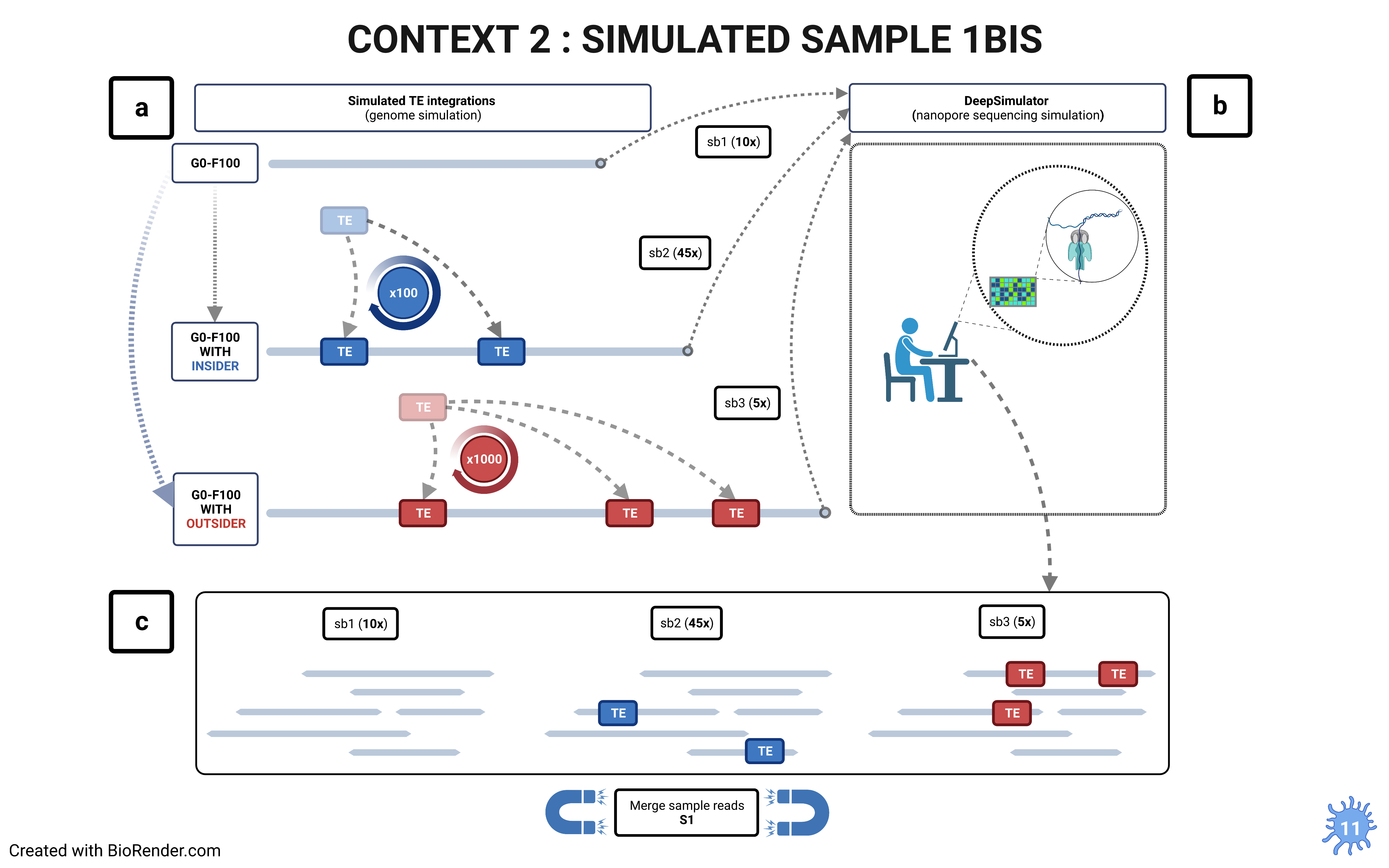
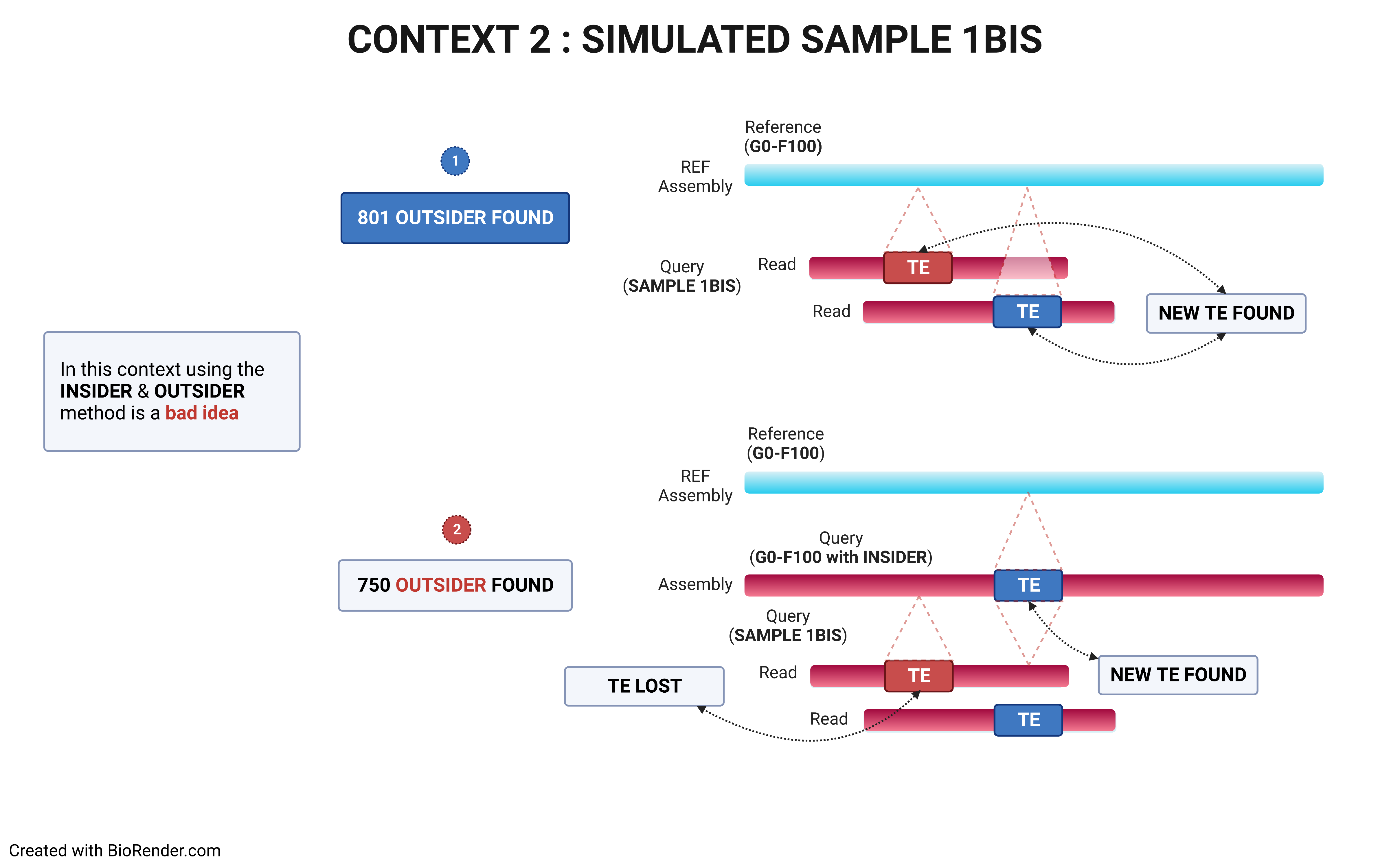
- strategies
- Citation & Licence
Transposable Elements MOvement detection using LOng reads
TrEMOLO uses long reads, either directly or through their assembly, to detect:
- Global TE variations between two assembled genomes
- Populational/somatic variation in TE insertion/deletion
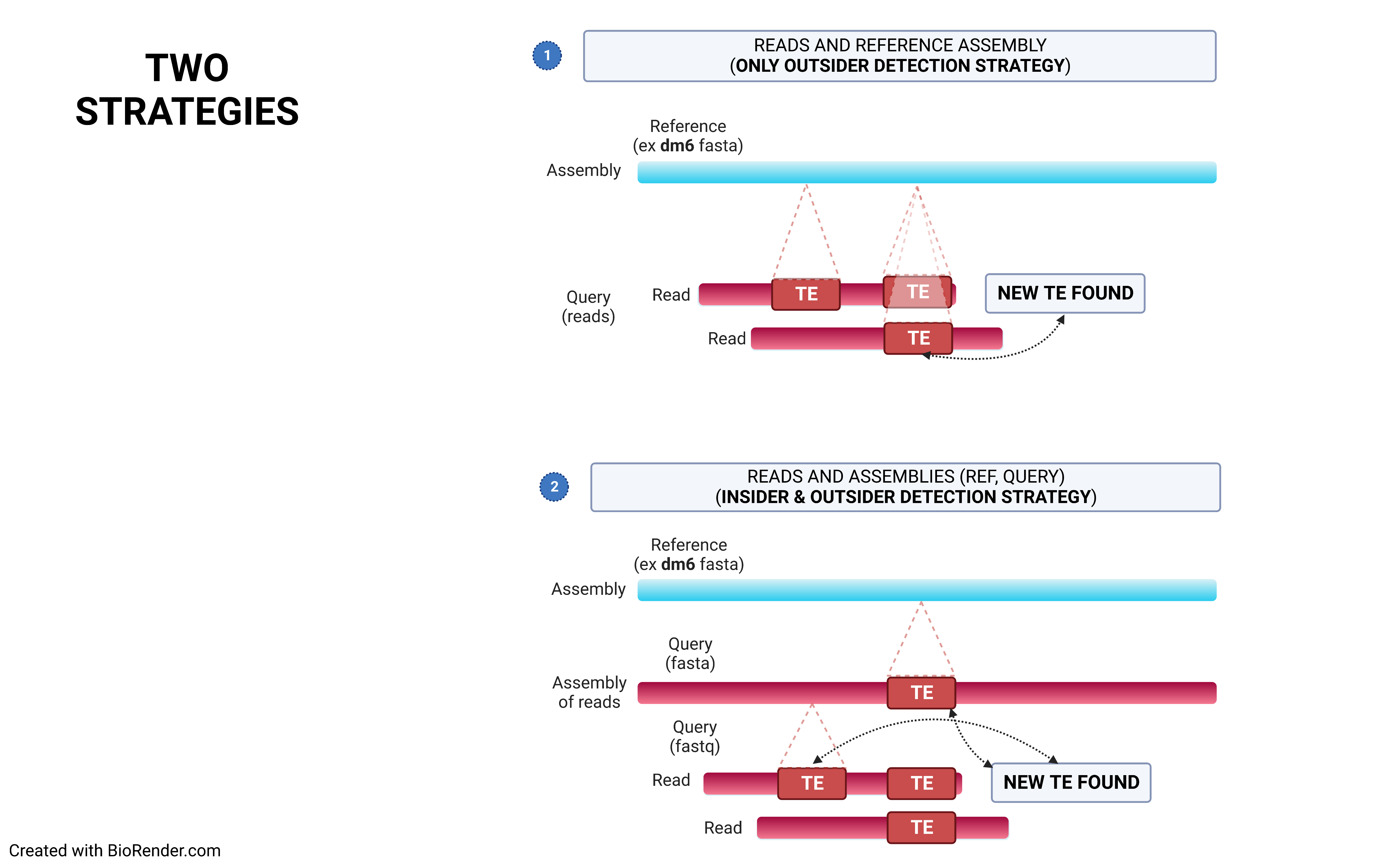
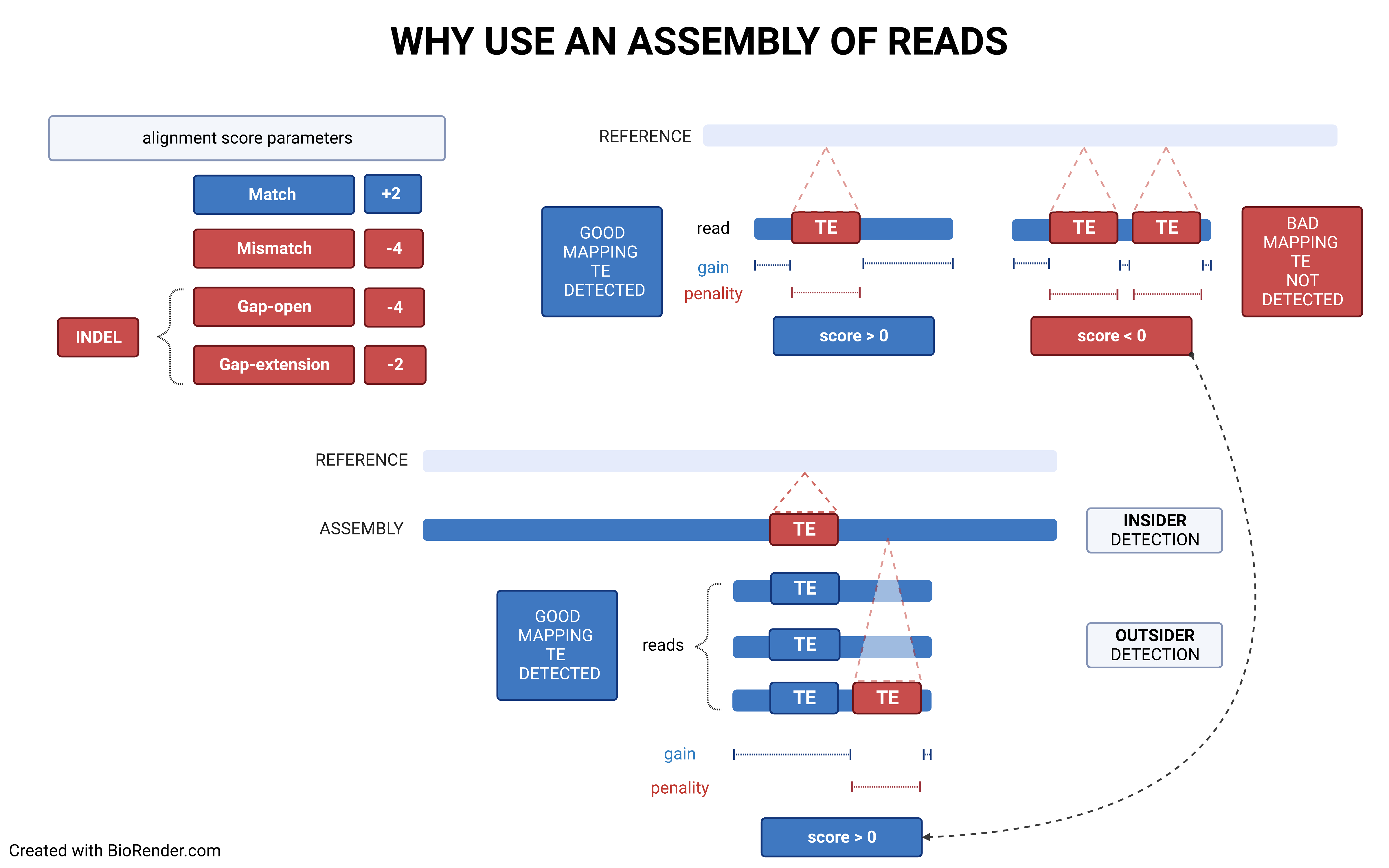
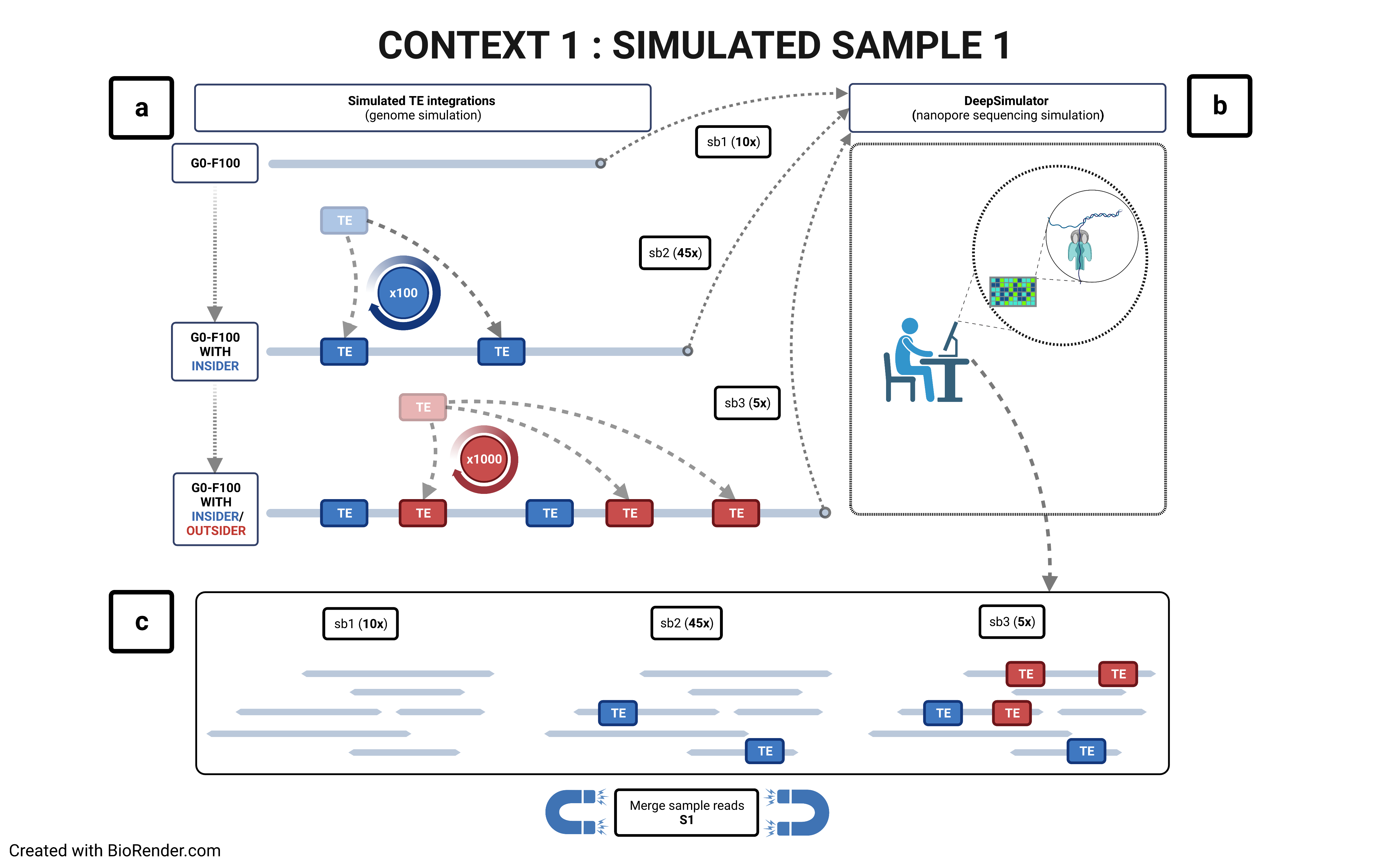
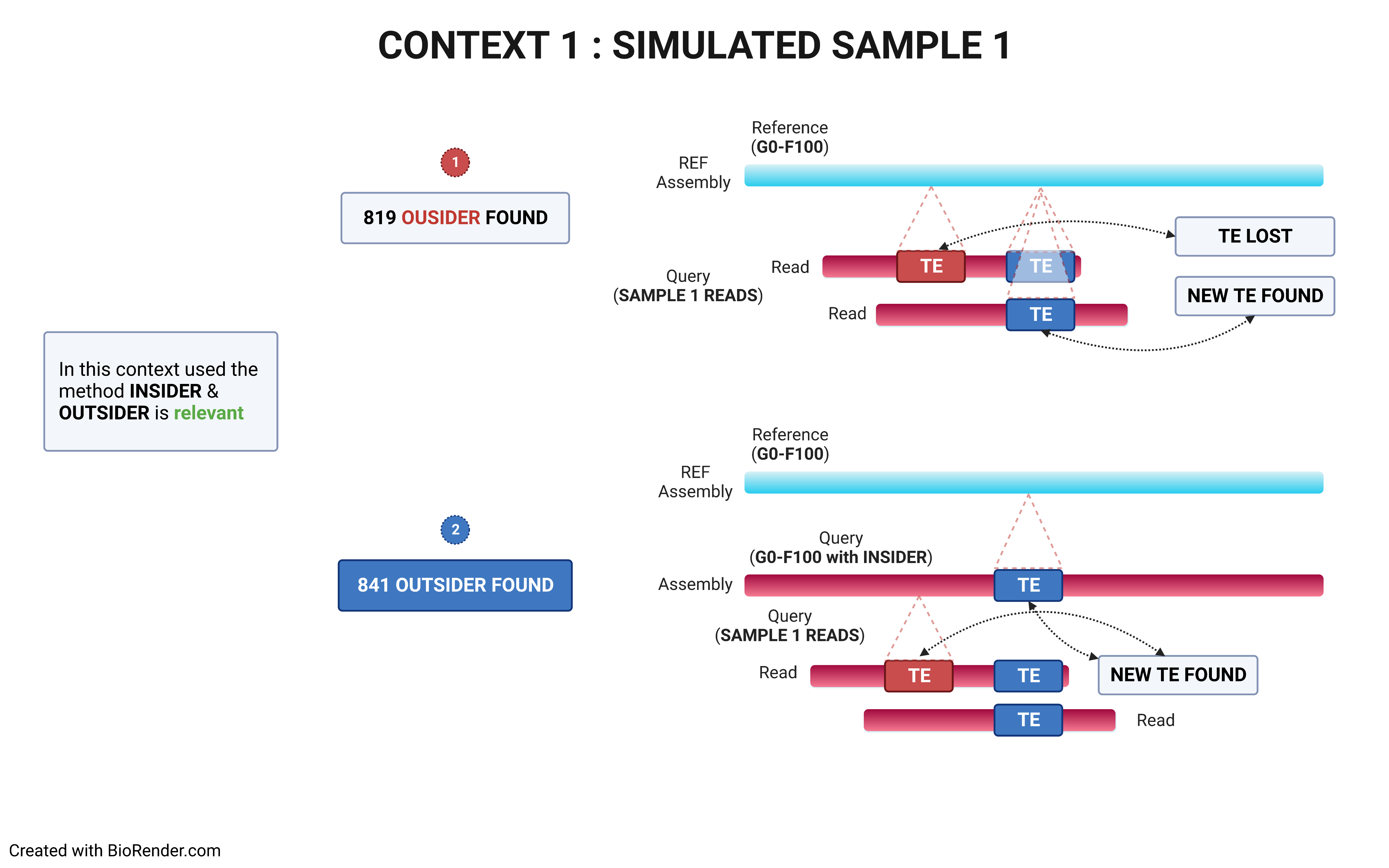
Using a reference genome and an assembled one (preferentially using long contigs or even better a chrosomome-scale assembly), TrEMOLO will extract the insiders, i.e. variant transposable elements (TEs) present globally in the assembly, and tag them. Indeed, assemblers will provide the most frequent haplotype at each locus, and thus an assembly represent just the "consensus" of all haplotypes present at each locus. You will obtain a set of files with the location of these variable insertions and deletions.
Through remapping of reads that have been used to assemble the genome of interest, TrEMOLO will identify the populational variations (and even somatic ones) within the initial dataset of reads, and thus of DNA/individuals sampled. These variant TEs are the outsiders, present only in a part of the population or cells. In the same way as for insiders, you will obtain a set of files with the location of these variable insertions and deletions.
Version 2.5.4
-
Update : Packages R Updated
bookdown- 0.38rmarkdown- 2.26
-
Change : Modifications in
rules.snkFiles- The
FIND_SV_ON_REF,FIND_TE_ON_REFrules have been replaced byLIFT_OFF.
- The
-
Add : New Parameters in
config.yamlfor INSIDERMINIMAP2:PRESET_OPTION: 'asm5'OPTION: '--cs'
-
Add : New Modules
- Scatter Frequency - Provides analysis of frequency variations across multiple generations.
- Analysis TE BLAST - Analyzes transposable elements using BLAST.
-
In INSIDER_VARIANT mode, TE annotation on the REFERENCE (parameter INTEGRATE_TE_TO_GENOME) is suboptimal. Some TEs might not be annotated on the reference.
-
Difficulty in identifying the true positives concerning clipped insertions (SOFT, HARD)
Comprehensive TE Analysis
In our upcoming release, we will be expanding our analysis capabilities to include a comprehensive examination of Transposable Elements (TEs) within both reads and genomes. This enhancement will go beyond merely identifying INDELs to encompass a full spectrum analysis of TEs.
Numerous tools are used by TrEMOLO. We recommand to use the Singularity installation to be sure to have all of them in the good configurations and versions.
- For both approaches
- Python 3.6+
- For Global variation tool
- BLAST 2.2+
- Bedtools 2.27.1 v2
- Assemblytics or
- RaGOO
- Liftoff
- For Populational variation tool
- Snakemake 5.5.2+
- Minimap2 2.24+
- Samtools 1.9 and (1.15.1 optional)
- svim 1.4.2
- Sniffles 1.0.12
- Python libs
- Perl v5.26.2+
- For report
- Others
- nodejs
Once the requirements fullfilled, just git clone
git clone https://github.com/DrosophilaGenomeEvolution/TrEMOLO.gitSingularity installation Debian/Ubuntu with package
A Singularity container (version 3.10.0+ required) is available with all tools compiled in. The Singularity file provided in this repo and can be compiled as such:
sudo singularity build TrEMOLO.simg TrEMOLO/SingularityYOU MUST BE ROOT for compiling
Alternatively, you can download a pre-compiled Singularity container from the following link:
Download TrEMOLO Singularity Container
Test TrEMOLO with singularity
singularity exec TrEMOLO.simg snakemake --snakefile TrEMOLO/run.snk --configfile TrEMOLO/test/tmp_config.yml
#OR
singularity run TrEMOLO.simg snakemake --snakefile TrEMOLO/run.snk --configfile TrEMOLO/test/tmp_config.ymlThis option is disabled since Singularity Hub is for the moment in read-only. We are looking for a Singularity repo to ease the use.
TrEMOLO uses Snakemake to perform its analyses. You have then first to provide your parameters in a .yaml file (see an example in the config.yaml file). Parameters are :
# all path can be relative or absolute depending of your tree.
#It is advised to only use absolute path if you are not familiar with computer science or the importance of folder trees structure.
DATA:
GENOME: "/path/to/genome_file.fasta" #genome (fasta file) [required]
TE_DB: "/path/to/database_TE.fasta" #Database of TE (a fasta file) [required]
REFERENCE: "/path/to/reference_file.fasta" #reference genome (fasta file) only if INSIDER_VARIANT = True [optional]
SAMPLE: "/path/to/reads_file.fastq" #long reads (a fastq[.gz] file) only if OUTSIDER_VARIANT = True [optional]
#At least, provide either REFERENCE or SAMPLE. Both can be provided
WORK_DIRECTORY: "/path/to/directory" #name of output directory [optional, will be created as 'TrEMOLO_OUTPUT']
#At least, you must provide either the reference file, or the fastq file or both
CHOICE:
PIPELINE:
OUTSIDER_VARIANT: True # outsiders, TE not in the assembly - population variation
INSIDER_VARIANT: True # insiders, TE in the assembly
REPORT: True # for getting a report.html file with graphics
OUTSIDER_VARIANT:
CALL_SV: "sniffles" # possibilities for SV tools: sniffles
INTEGRATE_TE_TO_GENOME: True # (True, False) Re-build the assembly with the OUTSIDER integrated in
CLIPPED_READS: False # (True, False) Processing of clipped reads (SOFT, HARD)
INSIDER_VARIANT:
DETECT_ALL_TE: False # detect ALL TE on genome (parameter GENOME) assembly not only new insertion. Warning! it may be take several hours on big genomes
INTERMEDIATE_FILE: True # Conserve the intermediate analyses files to process them latter.
PARAMS:
THREADS: 8 #number of threads for some task
OUTSIDER_VARIANT:
MINIMAP2:
PRESET_OPTION: 'map-ont' # minimap2 option is map-ont by default (map-pb, map-ont)
OPTION: '' # more option of minimap2 can be specified here
SAMTOOLS_VIEW:
PRESET_OPTION: ''
SAMTOOLS_SORT:
PRESET_OPTION: ''
SAMTOOLS_CALLMD:
PRESET_OPTION: ''
TSD:
SIZE_FLANK: 15 # flanking sequence size for calculation of TSD; put value > 4
TE_DETECTION:
CHROM_KEEP: "." # regular expresion for chromosome filtering; for instance for Drosophila "2L,2R,3[RL],X" ; Put "." to keep all chromosome
GET_SEQ_REPORT_OPTION: "-m 30" #sequence recovery file in the vcf
PARS_BLN_OPTION: "--min-size-percent 80 --min-pident 80 -k 'INS|DEL'" # option for TrEMOLO/lib/python/parse_blast_main.py - don't put -c option
INSIDER_VARIANT:
PARS_BLN_OPTION: "--min-size-percent 80 --min-pident 80" # parameters for validation of insiders
MINIMAP2:
PRESET_OPTION: 'asm5' # minimap2 preset option is asm5 by default (asm5, asm10, asm20 etc)
OPTION: '--cs'
The main parameters are:
GENOME: Assembly of the sample of interest (or mix of samples), fasta file.TE_DB: A Multifasta file containing the canonical sequence of transposable elements. You can add also copy sequences but results will be more complex to interpretate.REFERENCE: Fasta file containing the reference genome of the species of interest.WORK_DIRECTORY: Directory that will contain the output files. If the directory does not exist it will be created; default value is TrEMOLO_OUTPUT.SAMPLE: File containing the reads used for the sample assembly.
You can use config_INSIDER.yaml for only INSIDER analysis or config_OUTSIDER.yaml for only OUTSIDER analysis.
To analyse INSIDER, only the REFERENCE , the GENOME, the TE_DB and the WORK_DIRECTORY are required.
To analyse OUTSIDER, only the SAMPLE , the GENOME, the TE_DB and the WORK_DIRECTORY are required.
snakemake --snakefile /path/to/TrEMOLO/run.snk --configfile /path/to/your_config.yamlFor running tests
snakemake --snakefile TrEMOLO/run.snk --configfile TrEMOLO/test/tmp_config.ymlHere is the structure of the output files obtained after running the pipeline.
WORK_DIRECTORY
├── params.yaml ##**Your config file
├── LIST_HEADER_DB_TE.csv ##** list of names assigned to TE in the TE database (Only if you have charactere "& ; / \ | ' : ! ? " in your TE database)
├── POSITION_ALL_TE.bed -> INSIDER/TE_DETECTION/POSITION_ALL_TE.bed ##**ALL TE ON GENOME NOT ONLY INSERTION (ONLY IF PARAMETER "DETECT_ALL_TE" is True),
├── POSITION_TE_INOUTSIDER.bed
├── POSITION_TE_INSIDER.bed
├── POSITION_TE_OUTSIDER.bed
├── POS_TE_INSIDER_ON_REF.bed -> INSIDER/TE_DETECTION/INSERTION_TE_ON_REF.bed ##**POSITION TE INSIDER ON REFRENCE GENOME
├── POS_TE_OUTSIDER_ON_REF.bed ##**POSITION TE OUTSIDER ON REFRENCE GENOME
├── POSITION_TE_OUTSIDER_IN_NEO_GENOME.bed ##**POSITION TE SEQUENCE ON BEST READS SUPPORT INTEGRATED IN GENOME
├── POSITION_TE_OUTSIDER_IN_PSEUDO_GENOME.bed ##**POSITION TE SEQUENCE ON TE DATABASE (with ID) INTEGRATED IN GENOME
├── VALUES_TSD_ALL_GROUP.csv
├── VALUES_TSD_GROUP_OUTSIDER.csv
├── VALUES_TSD_INSIDER_GROUP.csv
├── TE_INFOS.bed ##**FILE CONTENING ALL INFO OF TE INSERTION
├── DELETION_TE.bed -> INSIDER/TE_DETECTION/DELETION_TE.bed ##**TE DELETION POSTION ON GENOME
├── DELETION_TE_ON_REF.bed -> INSIDER/TE_DETECTION/DELETION_TE_ON_REF.bed ##**TE DELETION POSITION ON REFERENCE
├── SOFT_TE.bed -> OUTSIDER/TE_DETECTION/SOFT/SOFT_TE.bed ##**TE INSERTION FOUND IN SOFT READS
├── INSIDER ##**FOLDER CONTAINS FILES TRAITEMENT INSIDER
│ ├── FREQ_INSIDER
│ ├── TE_DETECTION
│ ├── TSD
│ │ └── TSD_TE.tsv
│ ├── TE_INSIDER_VR
│ └── VARIANT_CALLING
├── log ##**log file to check if you have any error
├── OUTSIDER
│ ├── ET_FIND_FA
│ │ ├── TE_REPORT_FOUND_TE_NAME.fasta
│ │ ├── TE_REPORT_FOUND_blood.fasta
│ │ └── TE_REPORT_FOUND_ZAM.fasta
...
│ ├── FREQUENCY
| | ├── FREQUENCY_TE_INS_PRECISE.fasta
│ │ └── FREQUENCY_TE_INS.tsv
│ ├── INSIDER_VR
│ ├── MAPPING ##**FOLDER CONTAINS FILES MAPPING ON GENOME
│ ├── MAPPING_TO_REF ##**FOLDER CONTAINS FILES MAPPING ON REFERENCE GENOME
│ ├── TE_DETECTION
│ │ └── MERGE_TE
│ ├── TSD
│ │ └── TSD_TE.tsv
│ ├── TrEMOLO_SV_TE
│ │ ├── INS
│ │ ├── HARD
│ │ └── SOFT
│ ├── TE_TOWARD_GENOME ##**FOLDER CONTAINS ALL THE READs ASSOCIATED WITH THE TE
│ │ ├── NEO_GENOME.fasta ##**GENOME CONTAINS TE OUTSIDER (the best sequence of svim/sniffles)
│ │ ├── PSEUDO_GENOME_TE_DB_ID.fasta ##**GENOME CONTAINS TE OUTSIDER (the sequence of database TE and the ID of svim/sniffles)
│ │ ├── TRUE_POSITION_TE_PSEUDO.bed ##**POSITION IN PSEUDO GENOME
│ │ ├── TRUE_POSITION_TE.fasta ##**SEQUENCE INTEGRATE IN PSEUDO GENOME
│ │ ├── TRUE_POSITION_TE_NEO.bed ##**POSITION IN NEO GENOME
│ │ └── TRUE_POSITION_TE_READS.fasta ##**SEQUENCE INTEGRATE IN NEO GENOME
│ └── VARIANT_CALLING ##**FOLDER CONTAINS FILES OF sniflles/svim
├── REPORT
│ ├── mini_report
│ └── report.html
├── SNAKE_USED
│ ├── Snakefile_insider.snk
└── └── Snakefile_outsider.snk
The most useful output files are :
- The html report in your_work_directory/REPORT/report.html with summary graphics, as shown here
The output file your_work_directory/TE_INFOS.bed gathers all the necessary information.
| chrom | start | end | TE|ID | strand | TSD | pident | psize_TE | SIZE_TE | NEW_POS | FREQ (%) | FREQ_OPTIMIZED (%) | SV_SIZE | ID_TrEMOLO | TYPE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2R_RaGOO_RaGOO | 16943971 | 16943972 | roo|svim.INS.175 | + | GTACA | 97.026 | 99.2 | 9006 | 16943978 | 28.5714 | 28.5714 | 9000 | TE_ID_OUTSIDER.94047.INS.107508.0 | INS |
| X_RaGOO_RaGOO | 21629415 | 21629416 | ZAM|Assemblytics_w_534 | - | CGCG | 98.6 | 90.5 | 8435 | 21629413 | 11.1111 | 10.0000 | 8000 | TE_ID_INSIDER.77237.Repeat_expansion.8 | Repeat_expansion |
chrom: chromosomestart: start position for the TEend: end position for the TETE|ID: TE name and ID in SV.vcf,SV_SOFT.vcf,HARD.fasta and SV_INS_CLUST.bed (for OUTSIDER) or assemblytics_out.Assemblytics_structural_variants.bed (for INSIDER)strand: strand of the TETSD: TSD SEQUENCEpident: percentage of identical matches with TEpsize_TE: percentage of size with TE in databaseSIZE_TE: TE sizeNEW_POS: position corrected with calculated TSD (only for OUTSIDER)FREQ: frequency, normalizedFREQ_WITH_CLIPPED: frequency with clipped read (OUTSIDER only)SV_SIZE: size of the structural variant (may be larger than the size of the TE)ID_TrEMOLO: TrEMOLO ID of the TETYPE: type of insertion can be HARD,SOFT (Warning : HARD, SOFT are often false positives),INS,INS_DEL... (INS_DEL is an insertion located on a deletion of the assembly)
Modules are crucial tools in post-processing for analyses. They enable the extraction and visualization of complex information in an intuitive and accessible manner. With these modules, users can gain a deep understanding of data by directly visualizing outcomes in various graphical formats, thereby facilitating the interpretation and utilization of research results or analyses.
The "Scatter Frequency TE Tremolo" module provides a crucial graphical tool for researchers studying the evolution of transposable element (TE) insertion frequencies across generations. It clearly visualizes the dynamics of these genomic elements, offering valuable insights into their behavior and potential for adaptation or evolutionary change within populations over extended periods. For more details, please consult the full documentation at this link.
This module enables the visualization of BLAST results concerning the newly detected transposable element insertions. It allows for the visual identification of specific structures such as LTR recombinations, transposable elements (TEs) inserted within other TEs, or more complex structures like clusters of TEs. This tool is crucial for genomic researchers aiming to deeply analyze the dynamics of TE insertions. For more details, please consult the full documentation at this link.
The choice of the right strategy depends on the context.
Mourdas MOHAMED.
This work is licensed under CC BY 4.0 for all docs and manuals. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
It is licencied under CeCill-C and GPLv3.
If you use TrEMOLO, please cite:
Mohamed, M.; Sabot, F.; Varoqui, M.; Mugat, B.; Audouin, K.; Pélisson, A.; Fiston-Lavier, A.-S. & Chambeyron S. TrEMOLO: accurate transposable element allele frequency estimation using long-read sequencing data combining assembly and mapping-based approaches. Genome Biol 24, 63 (2023). (https://doi.org/10.1186/s13059-023-02911-2)
Mohamed, M.; Dang, N. .-M.; Ogyama, Y.; Burlet, N.; Mugat, B.; Boulesteix, M.; Mérel, V.; Veber, P.; Salces-Ortiz, J.; Severac, D.; Pélisson, A.; Vieira, C.; Sabot, F.; Fablet, M.; Chambeyron, S. A Transposon Story: From TE Content to TE Dynamic Invasion of Drosophila Genomes Using the Single-Molecule Sequencing Technology from Oxford Nanopore. Cells 2020, 9, 1776. (https://www.mdpi.com/2073-4409/9/8/1776)
The data used in the paper are available here on DataSuds.