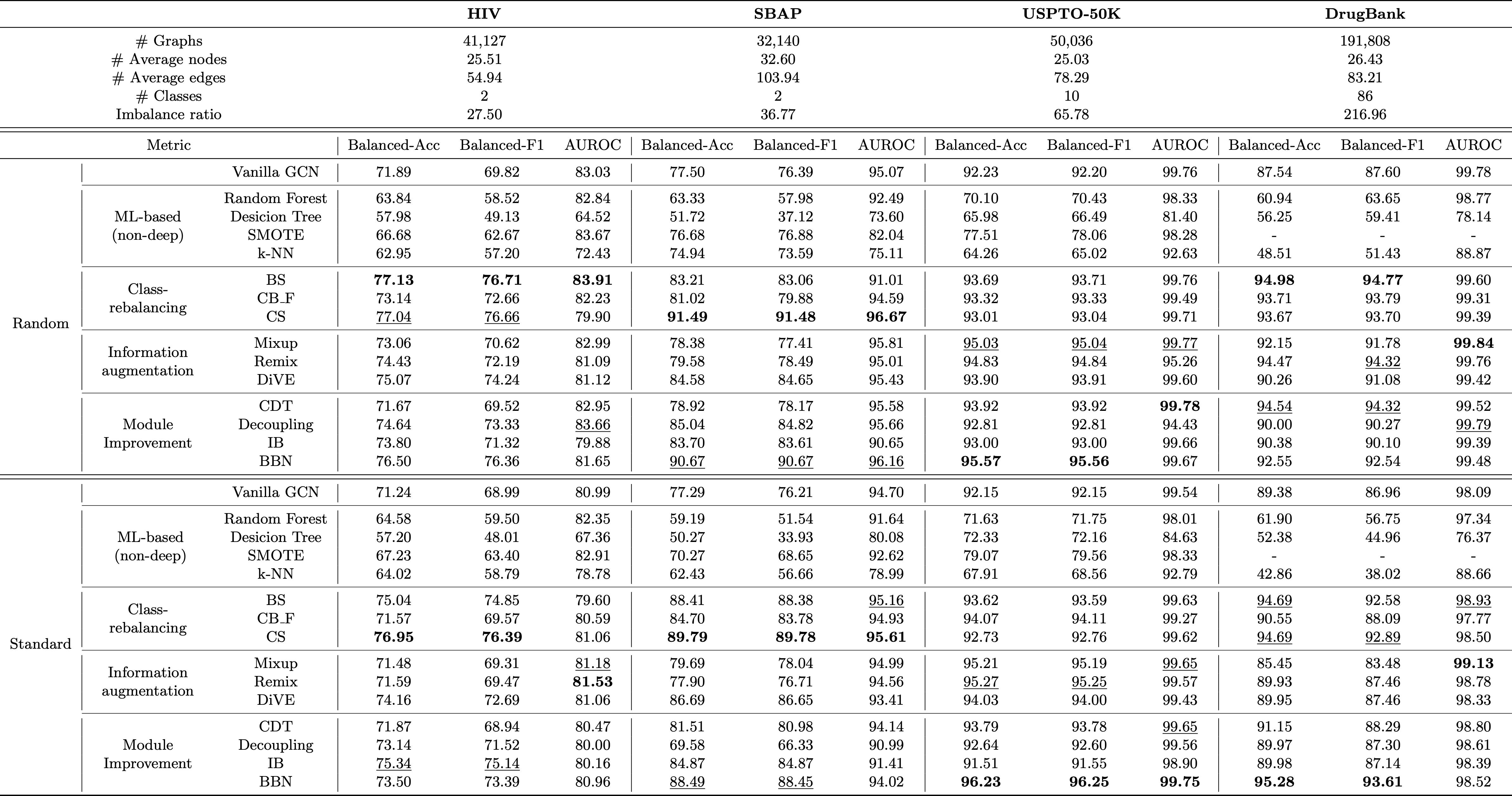
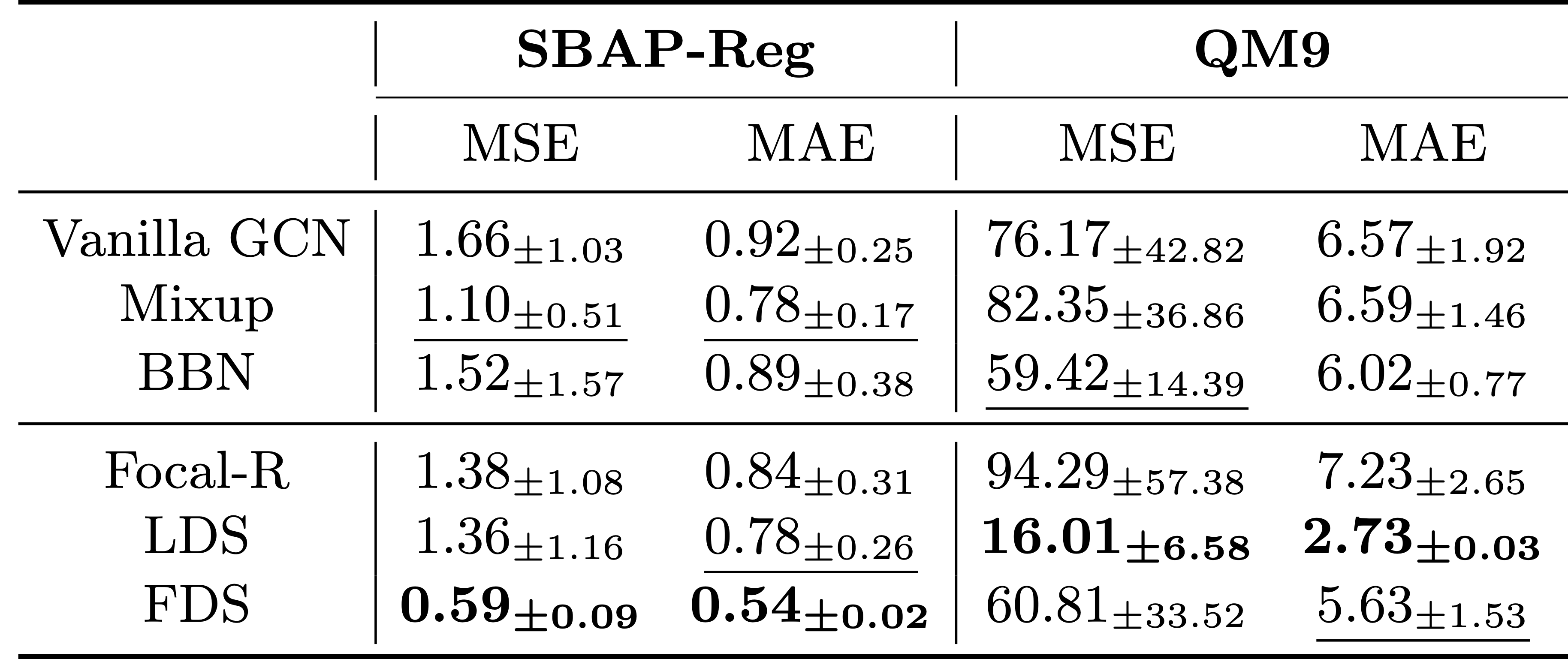
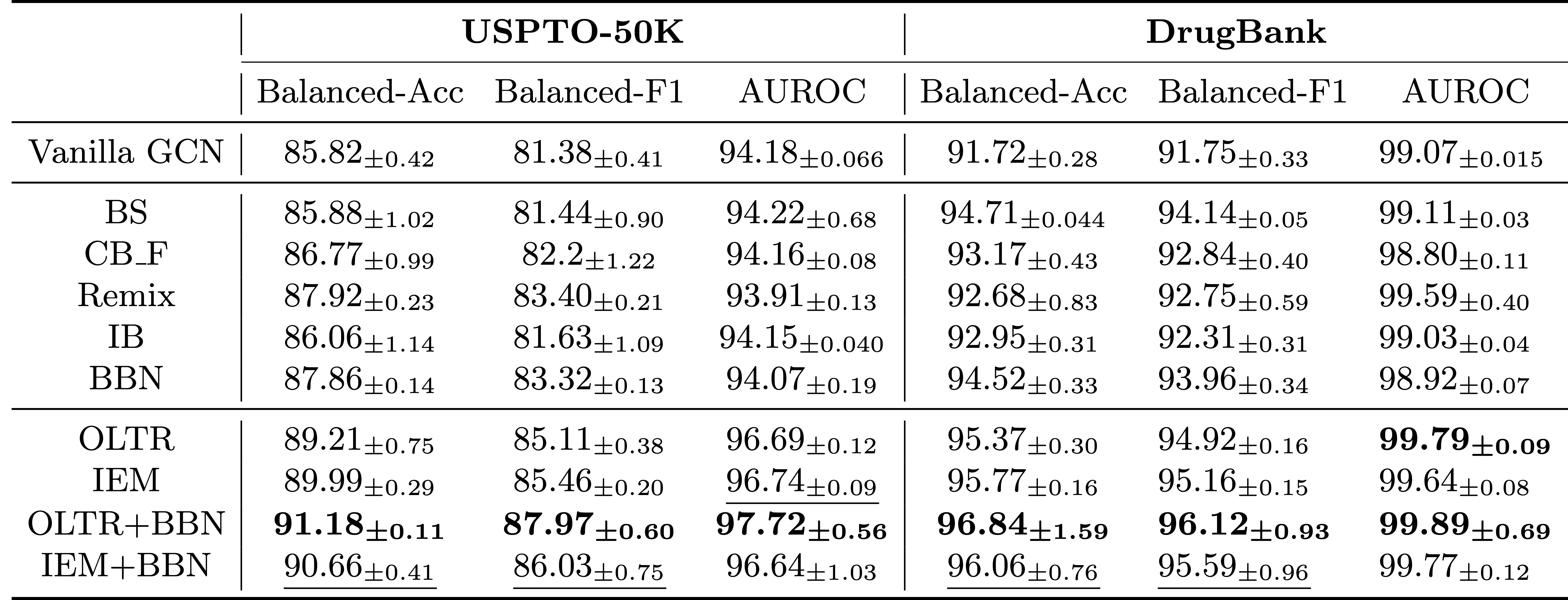
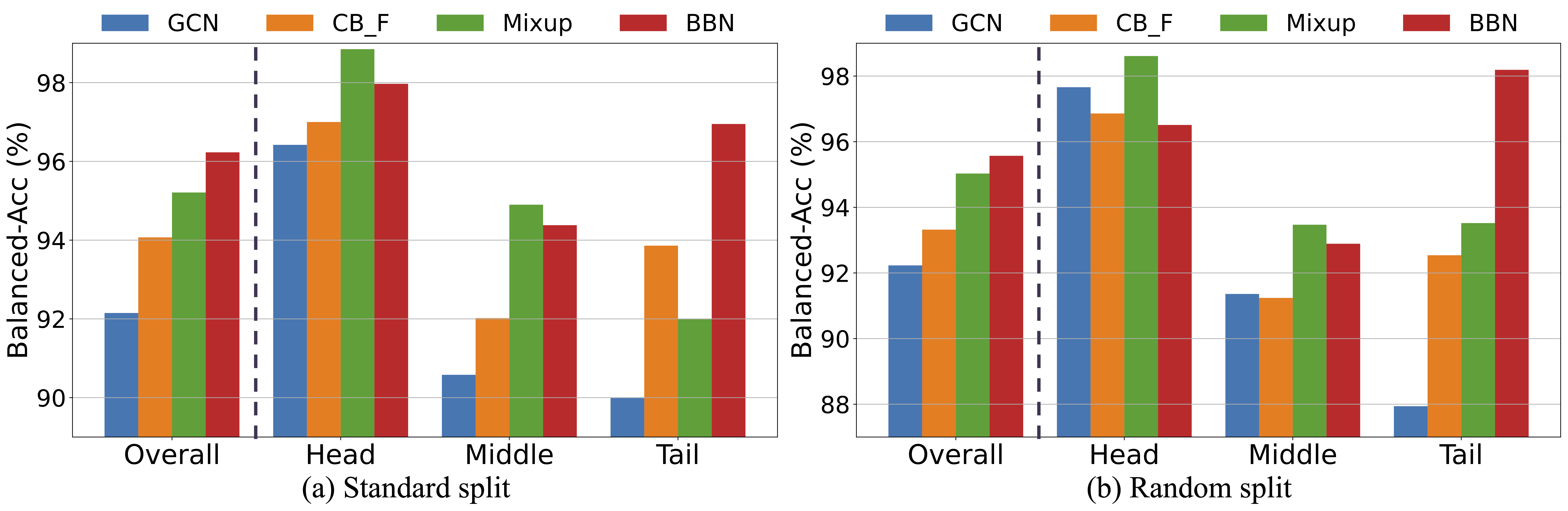
ImDrug is an open-source and systematic benchmark targeting deep imbalanced learning in AI-aided Drug Discovery. ImDrug features modularized components including formulation of learning setting and tasks, dataset curation, standardized evaluation, and baseline algorithms.
conda env create -f environment.yml
conda activate ImDrug
pip install git+https://github.com/bp-kelley/descriptastorusA task can be completely specified with an individual JSON file shown below. It updates the default configuration in /lib/config/default.py. Sample JSON files for producing the results in the paper can be found in /configs/.
{
"dataset": {
"drug_encoding": "DGL_GCN",
"protein_encoding": "Transformer",
"tier1_task": "multi_pred",
"tier2_task": "DTI",
"dataset_name": "SBAP",
"split":{
"method": "random",
"by_class": true
}
},
"baseline": "Remix_cls",
"test": {
"exp_id": "sbap_DGL_GCN_CrossEntropy_0_MLP_2022-06-09-00-10-53-662280"
},
"setting": {
"type": "LT Classification",
},
"use_gpu": true,
"save_step": 5,
"show_step": 5,
"valid_step": 1
}The 'dataset' entry in the JSON file specifies the dataset to be used, as well as the correponding data processing method with specifications such as featurization and data split. The configuration can be choosen as follows:
-
'drug_encoding' determines how the drugs/small-molecule compounds (if any) in the dataset will be featurized.
'drug_encoding': ["Morgan", "Pubchem’, "Daylight", "rdkit_2d_normalized", "ESPF", "CNN", "CNN_RNN", "Transformer", "MPNN", "ErG", "DGL_GCN", "DGL_NeuralFP", "DGL_AttentiveFP" "DGL_GIN_AttrMasking", "DGL_GIN_ContextPred"] -
'protein_encoding' determines how the proteins/large-molecules (if any) in the dataset will be featurized.
'protein_encoding': ["AAC", "PseudoAAC", "Conjoint_triad", "Quasi-seq", "ESPF", "CNN", "CNN_RNN", "Transformer"] -
'tier1_task' specifies the type of prediction problems.
'tier1_task': ["single_pred", "multi_pred"], both are applicable for hybrid prediction. -
'tier2_task' specifies the type of dataset and the prediction label.
'tier2_task': ["ADME", "TOX", "QM", "BioAct", "Yields", "DTI", "DDI", "Catalyst", "ReactType"] -
'dataset_name' specifies the dataset name.
'dataset_name': ["BBB_Martins", "Tox21", "HIV", "QM9", "USPTO-50K", "USPTO-Catalyst", "USPTO-1K-TPL", "USPTO-500-MT", "USPTO-Yields", "SBAP", "DrugBank"]- WARNING: Note that we keep the original format of "USPTO-500-MT" from Lu et al., for which we have confirmed with the authors that class 334 is missing. To use the dataset properly, one would need to make the class labels consecutive.
- WARNING: note that in principle, the yield of "USPTO-Yields" ranges from 0-1. However, the original copy of "USPTO-Yields" from TDC contains samples with negative yields or yields above 1, which we exclude in the current version.
-
'split.method' specifies the way to split the data, some of which rely on specific domain annotations such as scaffold and time splits.
'split.method': ["standard", "random", "scaffold", "time", "combination", "group", "open-random", "open-scaffold", "open-time", "open-combination", "open-group"], methods starting with "open-" are reserved for Open LT setting only.
The configuration of algorithms for imbalanced learning can be choosen as follows:
- For
LT ClassificationandImbalanced Classification:
'baseline': ["Default_cls", "BalancedSoftmax", "ClassBalanced", "CostSensitive", "InfluenceBalanced", "Mixup_cls", "Remix", "BBN_cls", "CDT", "Decoupling", "DiVE"] - For
Imbalanced Regression:
'baseline': ["Default_reg", "Mixup_reg", "Remix_reg", "BBN_reg", "Focal-R", "FDS", "LDS"] - For
Open LT:
'baseline': ["Default_cls", "BalancedSoftmax", "ClassBalanced", "InfluenceBalanced", "Remix", "BBN_cls", "OLTR", "IEM"]
Note that the suffix "cls" and "reg" indicate that the algorithm can be applied for both classification and regression tasks, respectively.
To run in Docker, go to ./script/docker. First download Miniconda3-latest-Linux-x86_64.sh and save it to ./common. Then run docker build . -t imdrug within that directory to build the Docker image tagged with the name imdrug. As an example, you can then run the container interactively with a bash shell with docker run --rm --runtime=nvidia -it -v [PATH_TO_ImDrug]:/root/code imdrug:latest /bin/bash.
Note that for the following examples, before running python3 script/test.py for inference, make sure to update cfg["test"]["exp_id"] in the JSON file to specify the experiment id and the saved model to be tested.
python3 script/train.py --config ./configs/single_pred/LT_Classification/baseline/HIV.json
python3 script/test.py --config ./configs/single_pred/LT_Classification/baseline/HIV.jsonpython3 script/train.py --config ./configs/single_pred/LT_Classification/information_augmentation/Remix/HIV.json
python3 script/test.py --config ./configs/single_pred/LT_Classification/information_augmentation/Remix/HIV.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Classification/baseline/SBAP.json
python3 script/test.py --config ./configs/multi_pred/LT_Classification/baseline/SBAP.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Classification/module_improvement/BBN/SBAP.json
python3 script/test.py --config ./configs/multi_pred/LT_Classification/module_improvement/BBN/SBAP.jsonpython3 script/train.py --config ./configs/single_pred/LT_Classification/baseline/USPTO-50k.json
python3 script/test.py --config ./configs/single_pred/LT_Classification/baseline/USPTO-50k.jsonpython3 script/train.py --config ./configs/single_pred/LT_Classification/class-re-balancing/BalancedSoftmaxCE/USPTO-50k.json
python3 script/test.py --config ./configs/single_pred/LT_Classification/class-re-balancing/BalancedSoftmaxCE/USPTO-50k.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Classification/baseline/USPTO-50k.json
python3 script/test.py --config ./configs/multi_pred/LT_Classification/baseline/USPTO-50k.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Classification/module_improvement/Decoupling/USPTO-50k.json
python3 script/test.py --config ./configs/multi_pred/LT_Classification/module_improvement/Decoupling/USPTO-50k.jsonpython3 script/train.py --config ./configs/single_pred/LT_Regression/baseline/QM9.json
python3 script/test.py --config ./configs/single_pred/LT_Regression/baseline/QM9.jsonpython3 script/train.py --config ./configs/single_pred/LT_Regression/LDS/QM9.json
python3 script/test.py --config ./configs/single_pred/LT_Regression/LDS/QM9.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Regression/baseline/SBAP.json
python3 script/test.py --config ./configs/multi_pred/LT_Regression/baseline/SBAP.jsonpython3 script/train.py --config ./configs/multi_pred/LT_Regression/FDS/SBAP.json
python3 script/test.py --config ./configs/multi_pred/LT_Regression/FDS/SBAP.jsonpython3 script/train.py --config ./configs/multi_pred/Open_LT/baseline/Drugbank.json
python3 script/test.py --config ./configs/multi_pred/Open_LT/baseline/Drugbank.jsonpython3 script/train.py --config ./configs/multi_pred/Open_LT/OLTR/Drugbank.json
python3 script/test.py --config ./configs/multi_pred/Open_LT/OLTR/Drugbank.jsonEach training process will generate a log (e.g., hiv_DGL_GCN_Transformer_MLP_2022-04-28-20-30.log) in ./output/{DATASET_NAME}/logs, and the models in ./output/{DATASET_NAME}/models/{EXP_ID}.
Note that before testing, you need to specify the training experiment id in cfg['test']['exp_id']. Each testing process will generate a log and a .pdf image of confusion matrix (e.g., BBB_Martins_Transformer_Transformer_MLP_2022-05-09-11-55.pdf) in ./output/{DATASET_NAME}/test.
To test trained models all at once, specify the "root_path" in ./test_all.py by the directory where all training logs are stored, i.e., root_path = ./output/{DATASET_NAME}/logs. Then run the following command line
python3 test_all.py ImDrug is hosted on Harvard Dataverse and Google Drive, both are available for manual download. Alternatively,
if you run any of the command lines in Running Examples, the necessary datasets will be automatically downloaded to the path specified in ./lib/config/default.py, from Harvard Dataverse.
Complete list of dataset_names:
- bbb_martins.tab
- hiv.tab
- tox21.tab
- qm9.tab
- sbap.tab
- drugbank.tab
- uspto_1k_TPL.tab
- uspto_500_MT.tab
- uspto_50k.tab
- uspto_catalyst.tab
- uspto_yields.tab
@article{li2022imdrug,
title={ImDrug: A Benchmark for Deep Imbalanced Learning in AI-aided Drug Discovery},
author={Li, Lanqing and Zeng, Liang and Gao, Ziqi and Yuan, Shen and Bian, Yatao and Wu, Bingzhe and Zhang, Hengtong and Lu, Chan and Yu, Yang and Liu, Wei and others},
journal={arXiv preprint arXiv:2209.07921},
year={2022}
}
ImDrug codebase is under the MIT license. The datasets are hosted on Harvard Dataverse under the CC0 1.0 license.
Reach us at imdrugbenchmark@gmail.com, lanqingli1993@gmail.com or open a GitHub issue.