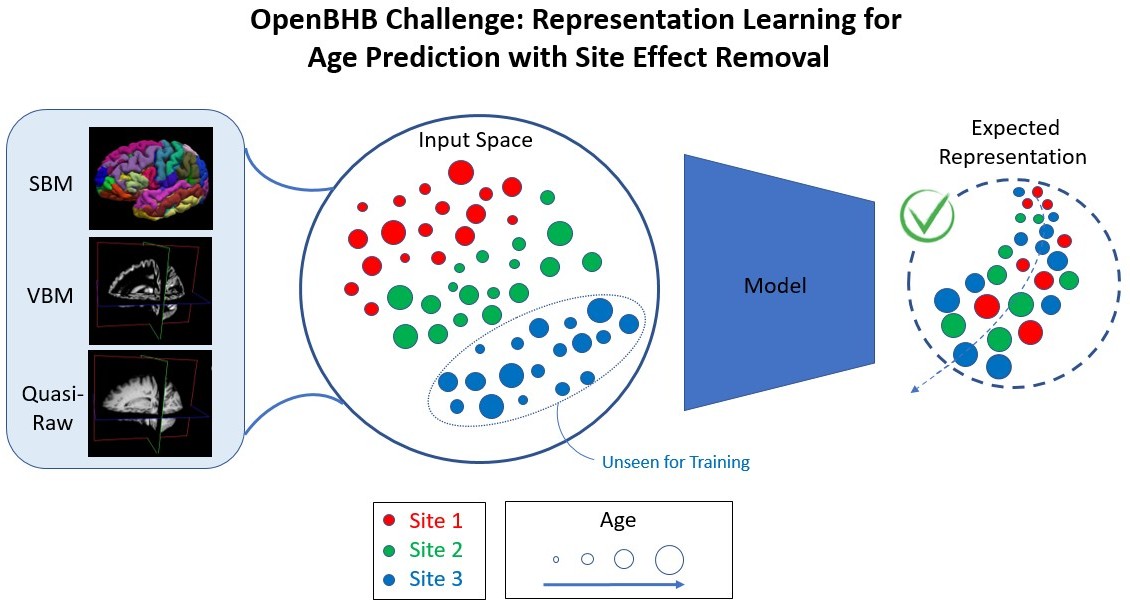
The challenge uses the openBHB dataset and aims to i) predict age from derived data from 3D T1 anatomical MRI while ii) removing site information from the learned representation. Thus, we aim to compare the capacity of proposed models to encode a relevant representation of the data (feature extraction and dimensionality reduction) that preserve the biological variability associated with age while removing the site-specific information. The algorithms submitted must output a low-dimension features vector (p < 10000). Derived data are composed of Quasi-Raw, VBM, and SBM.
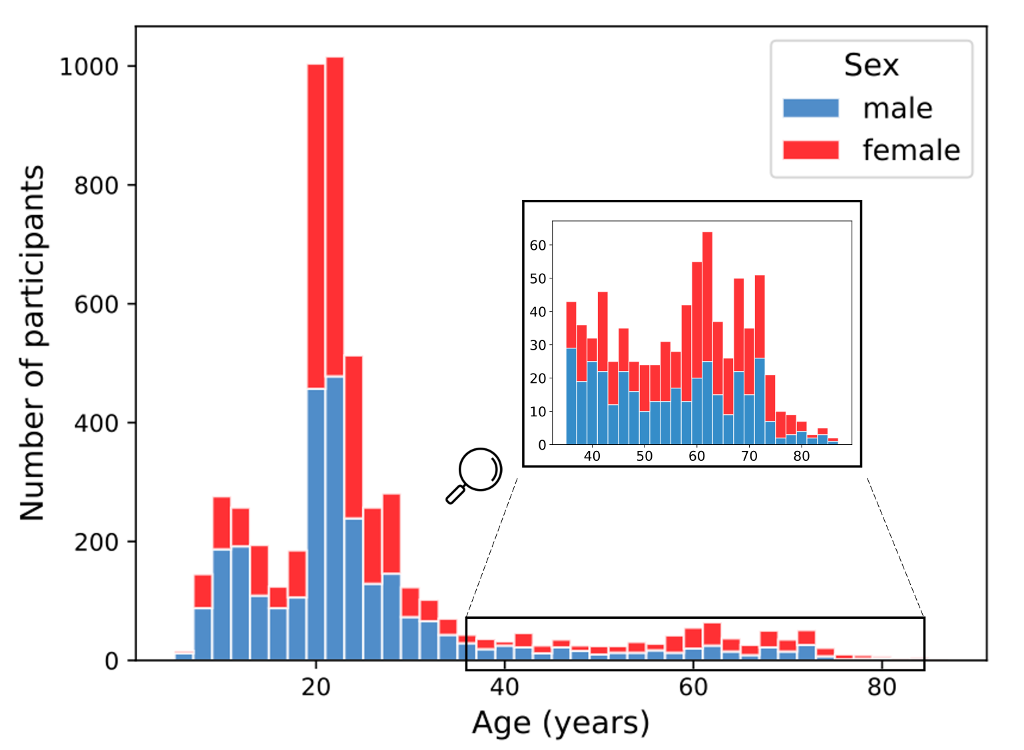
OpenBHB aggregates 10 publicly available datasets. Currently, openBHB is focused only on Healthy Controls (HC) since the main challenge consists in modeling the (normal) brain development by building a robust brain age predictor. OpenBHB contains
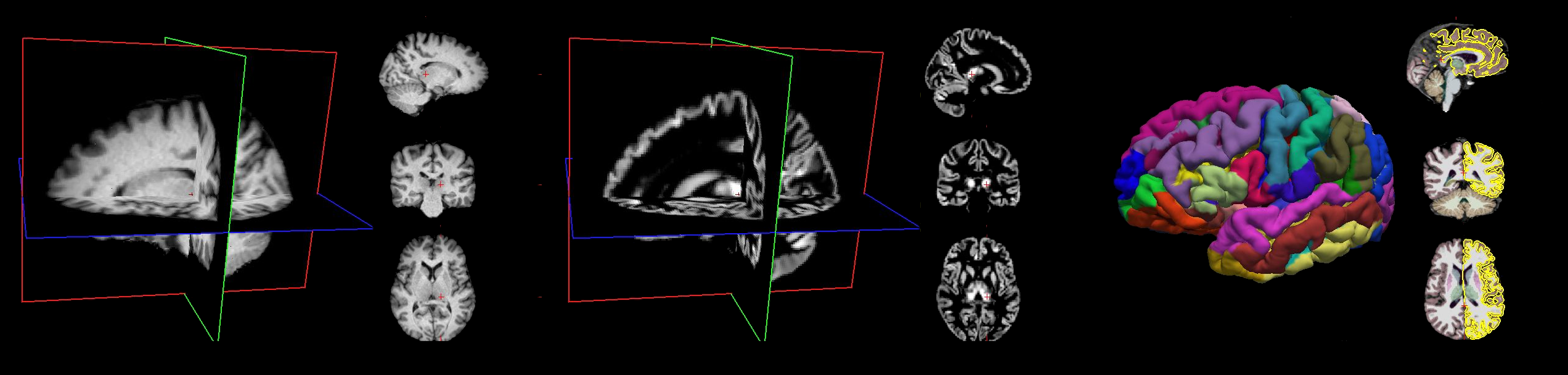
For the moment only features derived from T1w images are available comprising Quasi-Raw, CAT12 VBM, and FreeSurfer. All data are preprocessed uniformly including a semi-automatic Quality Controls (QC) guided with quality metrics.
The challenge will be carried out on the RAMP platform. It enables competition and collaboration on data-science problems, using the Python language. To start "hacking", a starting kit is available. It provides a simple working example which can be expanded to more advanced solutions.