This repository has been created to hold the containers used for the challenge 1 of the OpenEBench's implementation of LRGASP.
This repository updates the container registry used by OEB on release, allowing for several commits and pushes before a new version is released. 3 repositories are used:
Following the TCGA Benchmarking dockers example.
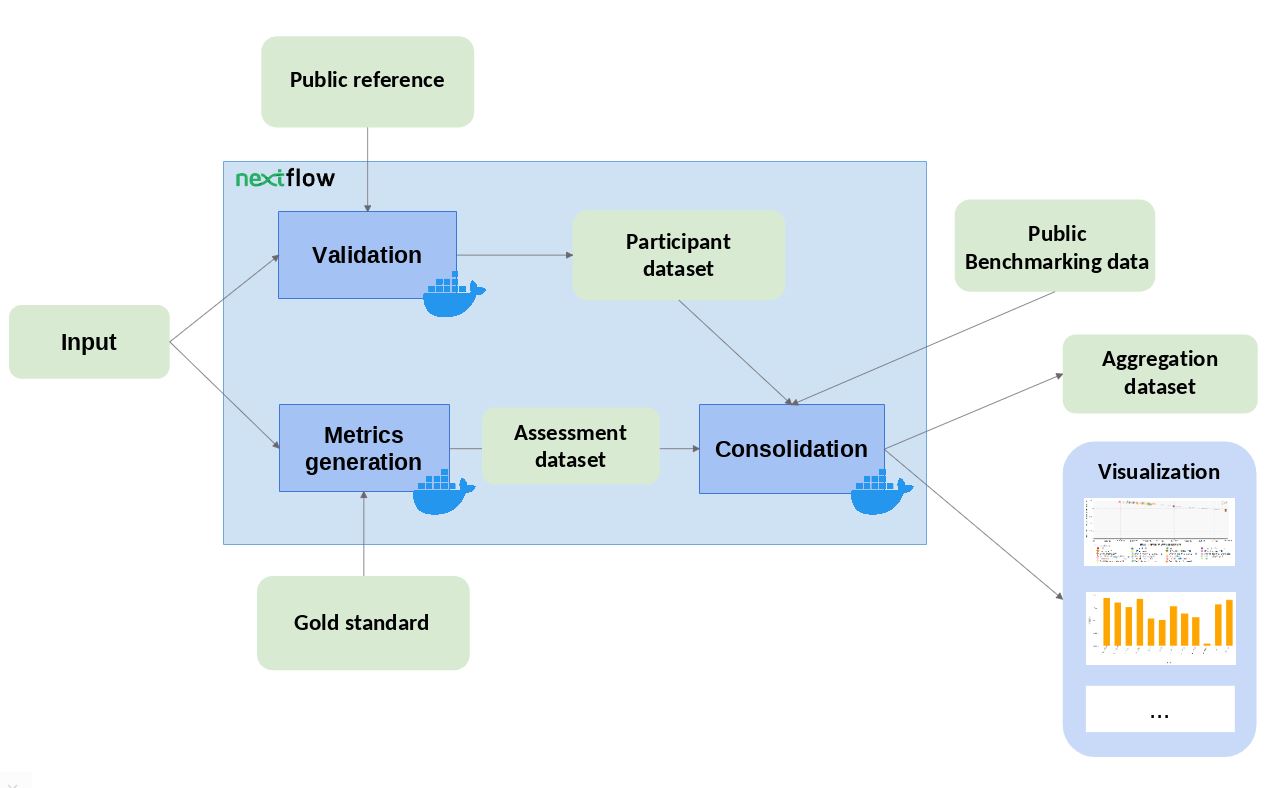
The structure of this repository follows the main structure as described in OEB specification page.
The specifics of this repository are described in the following subsections
- build.sh: This file, when run, will build the docker images for the validation, metrics and consolidation step. The tag provided (e.g.
1.0.0) needs to be the same as to when the pipeline is executed. - pipeline.bash: When run and given the proper tag id's, will execute the whole pipeline.
Contains the full data for LRGASP challenge 1, for WTC11.
This folder contains all the necessary files to run the pipeline.bash script. They belong to a real example. The IDs have been anonymised. To know how to used this data, please refer to the How to run section
This folder contains the scripts necessary for the validation step of the benchmark.
This folder contains the scripts necessary for the metrics step of the benchmark.
Sqanti3 is being used here to generate the reports.
This folder contains the scripts necessary for the consolidation step of the benchmark.
- Make sure you have installed docker
- Clone this repository
- Go to the root of this repository
- Download the missing files: The reference transcriptome and genome following the instructions here
- Run:
(If other versions are specified, please change it for the execution of pipeline.bash) This will build the docker images needed to run the three steps, with the 2.0.0 tag
sh build.sh 2.0.0Since we have not set up any parameters yet, it will print a usage statement:sh pipeline.bashUsage: pipeline.bash input_file TAG results_dir challenges When running, please ensure all the needed files are in the input_file, included the manifest with the filenames. The output dir must also be a full path - Re-run now, but adding the parameters specified in the previous bullet point. This will execute the whole pipeline, and all the results will be output to the results_dir. An example run with the example data:
sh pipeline.bash example_data/iso_detect_ref_input_example/input_user_files.tar.gz 2.0.0 Example/testresults "cdna_pacbio_ls_FSM cdna_pacbio_ls_SIRV cdna_pacbio_ls_NIC cdna_pacbio_ls_NNC cdna_pacbio_ls_ISM"
If you are interested in contributing to the repo, you can test changes to each of the steps individually by building a docker image for any of the steps (folders). An example with the validation step, after making the desired changes:
docker build -t "lrgasp_validation":"2.0.0" "lrgasp_validation"
docker run --rm -u $UID -v "example_data/iso_detect_ref_input_example":/app/input:rw -v "example_output":/app/output:rw lrgasp_validation:"2.0.0" \
-i /app/input/input_user_files.tar.gz -o /app/output/participant.json --challenges "$challenges" -m Please note:
- Each of the steps require a different set of inputs; for some information on what inputs they require, you can run the docker image without any arguments or take a look at the pipeline.bash script
- Please ensure that the outputs are still valid; depending on the steps, a set of outputs is required by the next step
The pipeline dies after some time executing. What is happening?
The "lrgasp_metrics" step executes sqanti3, a tool that requires at least 3 GB of ram memory allocation for this challenge. Please ensure that your docker is configured to allow at least 3 GB of ram memory for the containers. If after allocating more memory it keeps dying, please feel free to open a ticket and we will look into it.
If you want to contribute by improving the code, feel free to open PRs! Just remember to please update indirectly affected files - That accounts for:
- This README file: Anything that changes inputs or outputs on runtime should be taken into account here
- The example_data folder README file: Again, there is a comprehensive list of inputs and outputs there - Please update if modified.
- The requirements.txt: To modify them, we use
pip-compile, which takes into account the dependencies of your project dependencies to create a perfectly reproducible requirements file. To use, run this in the root of the repo:within your console or a set-up virtual environment. This will install pip-compile, which is used to transform requirements.in ("Raw" requirements files) into requirements.txt ("Dependency-thorough" requirement files). To use, just update thepip3 install pip-tools==6.13.0
requirements.infile and, in the same folder, runpython3 -m piptools compile
- all
sys.exit(-1)replaced withsys.exit(1).