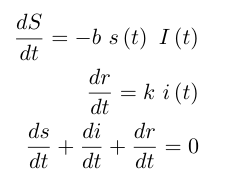Hackathon project submission repository for Pod 0.3.1 (Lazy Lobsters) Team 1
This project aims to use SciML to fit a differential equation to the COVID-19 Number-Of-Cases time series data. This would allow us to predict the number of cases in the future, to some degree of accuracy. This would be done separately per district or state, as per the availability of reliable data. The results would be displayed on an interactive graphical webpage.
The project also implements an interesting use case of the prediction model: a mobile application that is a personal scheduler/calendar. The user can enter where they are headed to in the next few days, and the application would warn them if those regions are predicted to have a high number of cases then.
Data sourced from https://api.covid19india.org/.
- Server code is present in
Serverdirectory - Datasets and the preprocessing script used are present in
Datasetdirectory - Trained models are in
BSONformat in theServer/datadirectory - Source of react native application is present in the
Application/directory with the build files (APK et al) inApplication/distdirectory
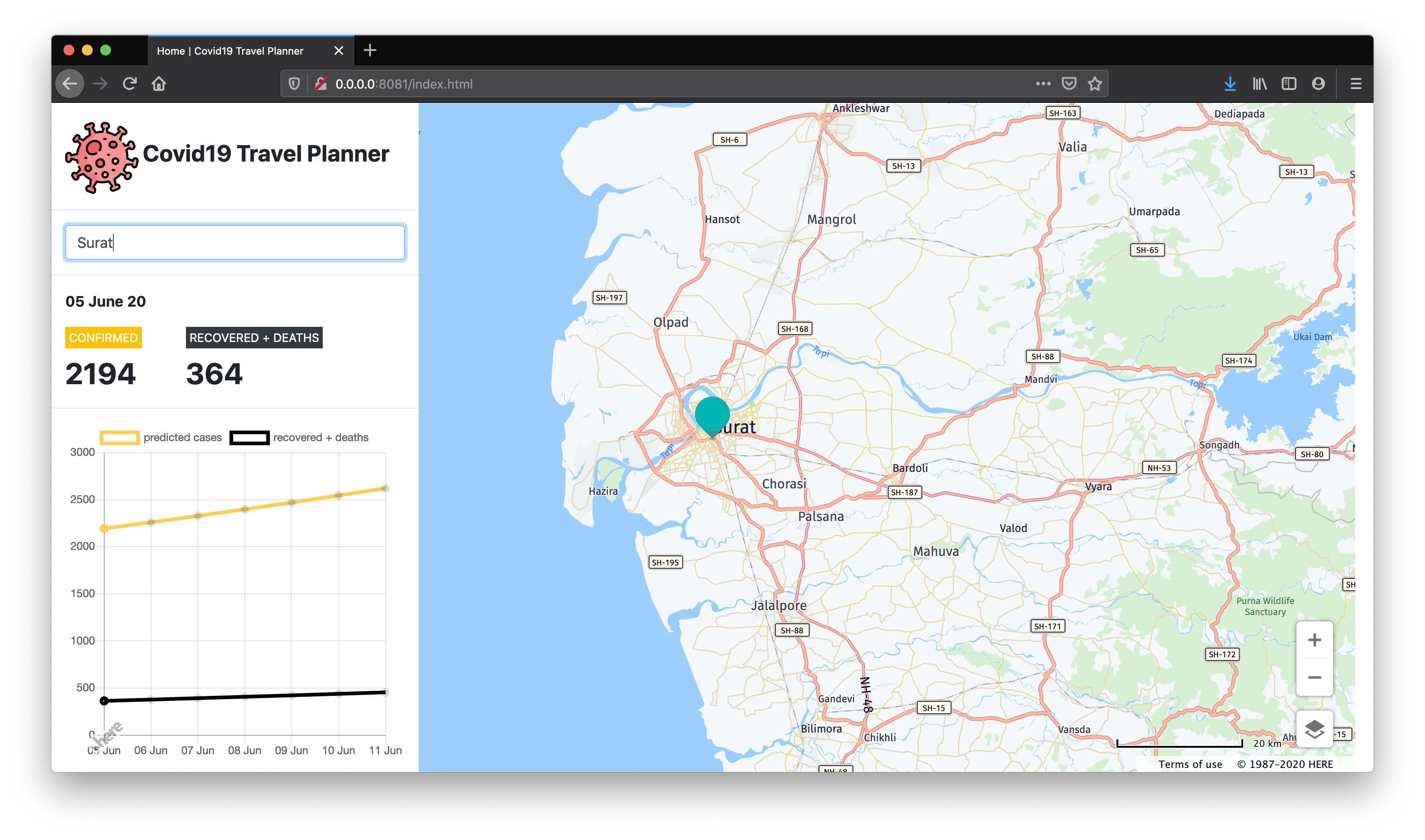
Web Application Homepage
- Julia : For the backend
- React Native : For the Application
- DiffEqFlux : For training district wise models
- React Native
- Expo
- Clone repository
git clone https://github.com/MLH-Fellowship/0.3.1-team1-COVIDTravelPlanner && cd 0.3.1-team1-COVIDTravelPlanner
- Install the server dependencies
julia --project=./Server -e "using Pkg; Pkg.instantiate()"
- Start the server
julia --project=./Server Server/server.jl
- Change into the
mobile-apprepository
cd mobile-app
- Install dependencies
npm install
- Run the Expo development server
npm start
- Follow the instructions on screen
We provide the code for training the neural network models and running inference using them in the Covid19Modelling module inside Server/lib. First we need to install the
dependencies using
julia --project=./Server -e "using Pkg; Pkg.instantiate()"
Our models need to be retrained daily / whenever a new data point is available. To train the model on a single district run the following code:
include("Server/lib/Covid19Modelling.jl") # Import the module
# The dataset needs to be updated on a daily basis
Covid19Modelling.train_named_locations("Dataset/final.csv", [<Names of Districts>], [<Names of States>], retrain = true, each_epochs = 100)If all the models need to be retrained, simply change the last line above to the following
Covid19Modelling.train_alll_districts("Dataset/final.csv", retrain = true, each_epochs = 100)Once the models are trained to obtain the predictions for the next 7 days, we do the following:
# Gives the predictions for the next 7 days
Covid19Modelling.prediction(<Name of state>, <Name of District>, [1, 2, 3, 4, 5, 6, 7])
# Gives the predictions for the 2nd, 4th and 6th day from today
Covid19Modelling.prediction(<Name of state>, <Name of District>, [2, 4, 6])
# Gives the predictions for day after tomorrow
Covid19Modelling.prediction(<Name of state>, <Name of District>, 2)Basically, you can get the predictions for any day in the next week. The function returns the total active cases and the total resolved cases (recovered + deceased)
We model our Differential Equations as follows using the DifferentialEquations.jl packages
We draw inspiration from the previous works [1] and [2]. We use a multilayer perceptron to parameterize the differential equations and using backpropogation through the solver we are able to learn the parameters in an end-to-end manner. To learn to predict the case counts we use minimize the weighted squared difference between the log of the case numbers.
[1] Rackauckas, Christopher, et al. "Universal Differential Equations for Scientific Machine Learning." arXiv preprint arXiv:2001.04385 (2020).
[2] Dandekar, Raj, and George Barbastathis. "Quantifying the effect of quarantine control in Covid-19 infectious spread using machine learning." medRxiv (2020).