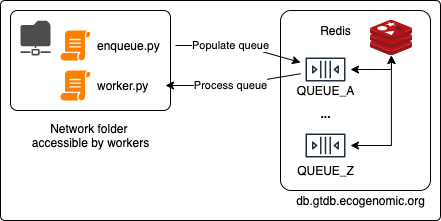
This repository is a template for distributing and orchestrating jobs across the ACE cluster.
Clone this repository into a new directory that is accessible by all worker nodes in the cluster (or, make note to copy it later).
git clone git@github.com:Ecogenomics/ace-cluster-orchestrator.gitIt is strongly recommended that you do all of this inside a new conda environment, or Python virtualenv.
The orchestrator uses Redis Queue (rq) and Redis-py (redis).
python -m pip install -r requirements.txtThis file will contain the logic to generate each of the jobs that will be run on the cluster.
It will require the Redis password to db.gtdb.ecogenomic.org (this can be found on BitWarden).
This is a trivial example, but to get started you will need to change the REDIS_PASS and QUEUE_NAME variables
at the beginning of the script.
The worker script will need to be importable by the enqueue.py file and will likely sit in the same directory.
It is going to be most useful to use subprocess here to spawn third party programs.
Ensure that you have uploaded the enqueue.py and worker.py files to the network location if you were editing them
on your local machine.
Simply run the enqueue.py file.
If you were using a conda environment, or Python virtualenv then activate it now.
Afterwards, change to the directory where enqueue.py and worker.py are located.
conda activate <env_name>
cd <path_to_repository>Activate the worker, ensuring that you have substituted the following variables:
REDIS_PASSfor the Redis password (note: the colon before it is required).QUEUE_NAMEfor the queue name defined inenqueue.py.WORKER_NAMEfor a human-readable name for the worker.
rq worker --name WORKER_NAME --burst --url "redis://:REDIS_PASS@db.gtdb.ecogenomic.org" "QUEUE_NAME"Note: --burst will terminate the worker as soon as QUEUE_NAME is empty.
Edit the Dockerfile and replace REDIS_PASS with the Redis password (keeping the colon in front of REDIS_PASS)
On your local machine (or any server on the UQ VPN), run the following docker command inside this directory:
docker run -p 9181:9181 -it $(docker build -q .)This will run rq-dashboard on port 9181: http://0.0.0.0:9181/
When running RQ with many hundreds of thousands of jobs, it is often best to not return values. This is because RQ will
store all results in the Redis database (as defined by the result_ttl). In that case, it's best to
write the results to disk.
Since there is only one Redis database, the queue names must be unique. It's best to check what queues exist before creating new ones.
Since this will often distribute jobs across the entire cluster, ensure that you are lowering
the priority of your workers. This can be done by putting os.nice(10) at the start of the worker code.
Note that this only affects CPU priority, if you require a lot of RAM this will not be fair.