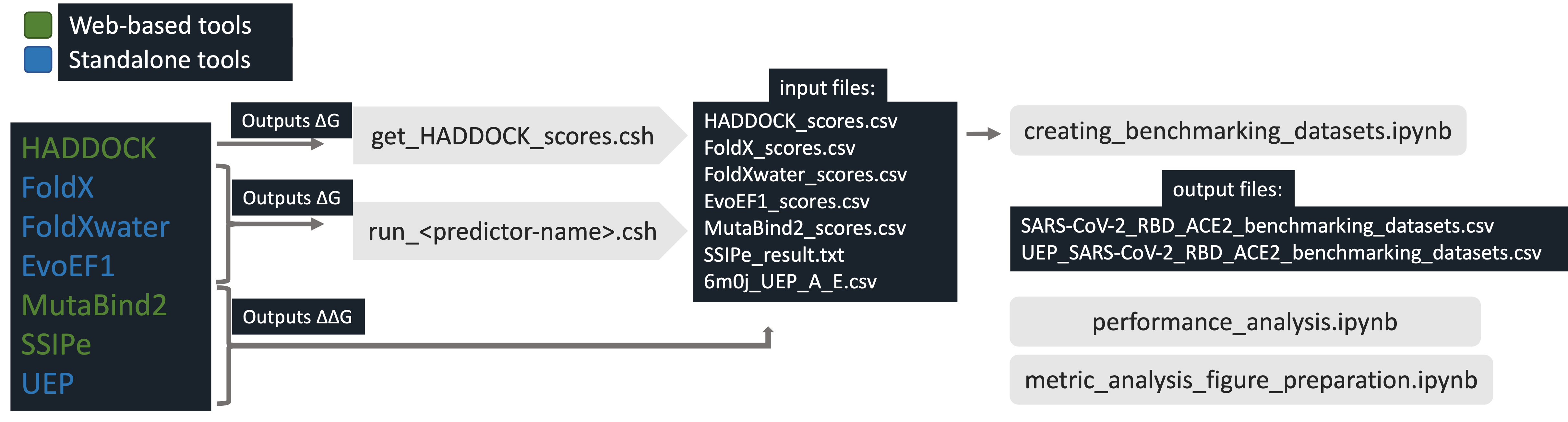
Benchmarking the structure-based binding energy predictors on human ACE2 - SARS-CoV-2 RBD binding data set.
Accurate prediction of binding energy change is a question of utmost importance in computational structural biology. There are many approaches developed to this aim to unveil the molecular mechanism underlying binding of two molecules and developing new therapeutic binders. In our study, we benchmarked six structure-based binding energy predictors; HADDOCK, FoldX, EvoEF1, MutaBind2, SSIPe, and UEP on the ACE2-RBD binding affinity dataset. Further we added FoldX with water option (FoldXwater) to contribute water mediated hydrogen bonds in the interface. Thus we benchmarked seven different approaches on the dataset that consist of 263 interface-point mutations. We presented prediction performance of the six approaches in this study. Additionally we assessed predictors' nature by using different metrics which related to amino acid change upon point mutation; van der Waals volume, hydrophobicity, flexibility, physicochemical property and position in the interface.
Presenting prediction performance of binding energy predictors is crucial to reveal what we can do with computationally inexpensive and fast methods in the case of future pandemics.
We released the study's files in a structured folder organization. Input files contain only binding energy outputs of predictors and experimental data sets. Raw data of six predictors were stored at Drive due to limitations of GitHub storage quotas. 263 point mutated ACE2-RBD protein complexes and corresponding binding affinity values are available on this link.
Assorted scripts related to the study were stored at the 'scripts' folder. Shell scripting was used for automatization of mutation generation and file manipulation -generating input files of study. Python notebooks were used for figure generation, performance and metric analyses. Generated figures and benchmarking dataset stored at 'output files' folder. The purpose of the scripts and the description of output files explained in detail at the below.
run_ scripts used for automatization of mutation generation of standalone tools. We generated FoldX and EvoEF1 models by using these scripts. HADDOCK, MutaBind2, and SSIPe models produced via their web server. Lastly, UEP is a standalone tool which is produce all possible mutations at once -no need automatization.
- run_FoldX.csh: Builds single amino acid mutations and computes binding affinity by using FoldX. (FoldX commands: Repair, BuildModel, AnalyseComplex).
- run_FoldXwater.csh: Builds single amino acid mutations and computes binding affinity by using FoldX with water option. (FoldX commands: Repair, BuildModel, AnalyseComplex with water)
- run_EvoEF1.csh: Builds single amino acid mutations and computes binding affinity by using EvoEF1. (EvoEF1 commands: RepairStructure, BuildMutant, ComputeBinding).
- get_HADDOCK_scores.csh: Used for grepping predicted HADDOCK binding energy scores from HADDOCK run files.
- pdb-tools.csh: Used to generate HADDOCK input structure since HADDOCK do not support multiple occupancy.
- creating_benchmarking_datasets.ipynb: Creates SARS_CoV_2_RBD_ACE2_benchmarking_dataset.csv and UEP_SARS_CoV_2_RBD_ACE2_benchmarking_dataset.csv files. These files are contain ∆∆G scores of predictors.
- performance_analysis.ipynb: Calculates prediction performance of predictors by using SARS_CoV_2_RBD_ACE2_benchmarking_dataset.csv and UEP_SARS_CoV_2_RBD_ACE2_benchmarking_dataset.csv files.
- metric_analyses_figure_preparation.ipynb: Builds metric analyses and creates figures of this study.
- ACE2_Experimental_dataset.csv: Experimental binding valus of ACE2 point mutations.
- RBD_Experimental_dataset.csv: Experimental binding valus of RBD point mutations.
- HADDOCK_scores.csv: Binding energy scores (∆G) of HADDOCK on 263 point mutations (+ wild type).
- FoldX_scores.csv: Binding energy scores (∆G) of FoldX on 263 point mutations, each mutations have their own wild type.
- FoldXwater_scores.csv: Binding energy scores (∆G) of FoldX with pdbWaters option on 263 point mutations, each mutation has its own wild type.
- EvoEF1_scores.csv: Binding energy scores (∆G) of EvoEF1 on 263 point mutations (+ wild type).
- MutaBind2_scores.csv: Binding energy change (∆∆G) scores of MutaBind2 on 263 point mutations.
- SSIPe_result.txt: Binding energy change (∆∆G) scores of SSIPe on 263 point mutations.
- 6m0j_UEP_A_E.csv: Binding energy change (∆∆G) scores of UEP.
- SARS-CoV-2-RBD_ACE2_benchmarking_dataset.csv: Main dataset of the study, contains ∆∆G scores of predictors on 263 point mutaions of SARS-CoV-2 RBD and human ACE2 protein complex (PDB ID: 6m0j).
- UEP_SARS-CoV-2-RBD_ACE2_benchmarking_dataset.csv: UEP calculates ∆∆G for highly-packed residues that have at least 2 non-covalent bond in the interface. So UEP dataset is a fraction of main dataset that represents core mutations.
- Figures.pdf: Metric related figures of the study.
git clone https://github.com/CSB-KaracaLab/ace2-rbd-point-mutation-benchmark