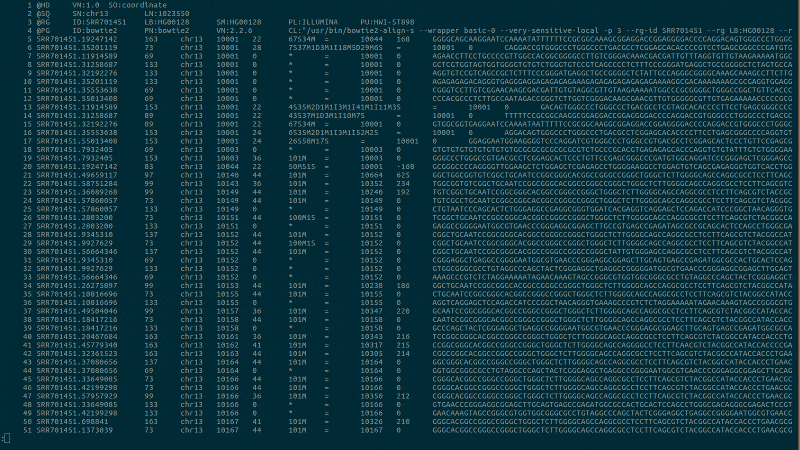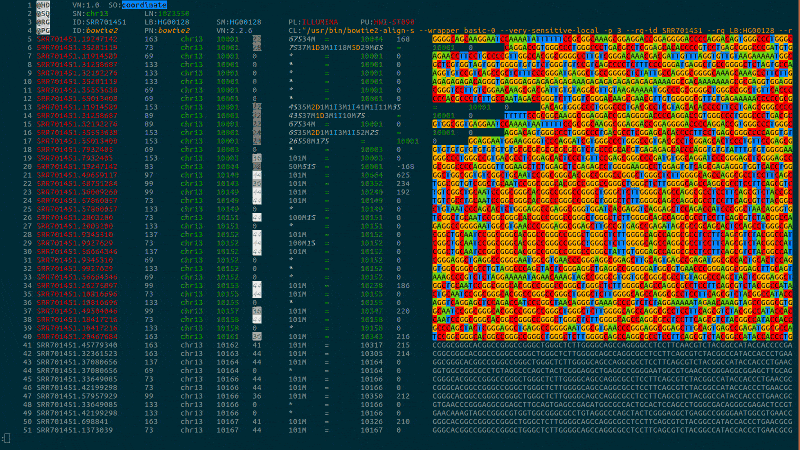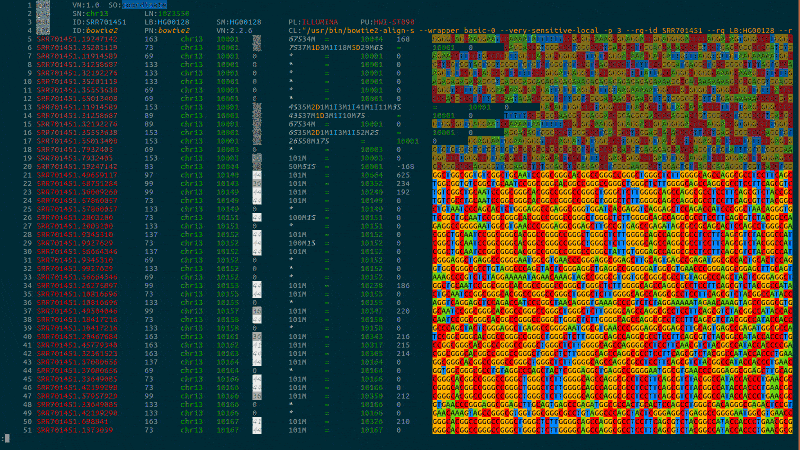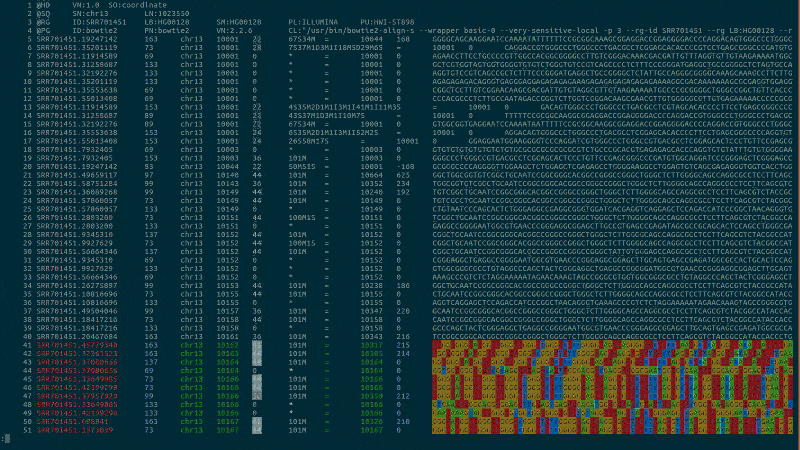
Syntax highlighting for computational biology to bring you intuitively close to your data.
Support in vim, less, sublime3 & gedit for .sam, .flagstat, .vcf, .fasta, .fastq, .faidx, .clustal, .pdb, .gtf, .bed files & more.
We'd like to know how bioSyntax can be developed to help your workflow. Tell us what you think! (~5m)
- Find out how you can contribute!
- Date: May 10 - May 11 (9:00 am - 5:00 pm PST)
- On-Site Location: Mozilla Vancouver, #209-163 W Hastings St, Vancouver, BC V6B 1H5, Canada (NOTE: No on-site parking available)
- Join us for the pre-Sprint Demo Call on Tuesday, May 8, 16:00 UTC to learn more about the project!
CONTACT:
- Project Lead: Alyssa Fegen (alyssafegen at alumni.ubc.ca); will be on-site
- Slack Channel (NOTE: Invite link expires on May 13; contact dev team for invites subsequently)
- Discord Server (NOTE: Invite link expires on May 13; contact dev team for invites subsequently)
bioSyntax integrates seamlessly with your text editor, recognizing data-type by the file extension or streamed into less.
To gain the most insight from your data, read our brief bioSyntax Manual.
samtools view -h NA19238.bam | sam-less -
This project is participating in the Mozilla Festival 2018, and as such, respects Mozilla's community participation guidelines. This serves as a guideline for the behaviour of all participants, remote or local, as well as project leads, facilitators, staff, volunteers, vendors, and anyone else involved in making the Mozilla Global Sprint possible.
Dev Team: Artem Babaian | Anicet Ebou | Alyssa Fegen | Jeffrey Kam | Gherman Novakovskiy | Jasper Wong
Contact us: info[AT]biosyntax.org
A hackseq17 Initiative.