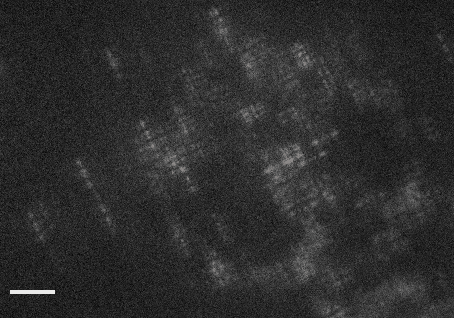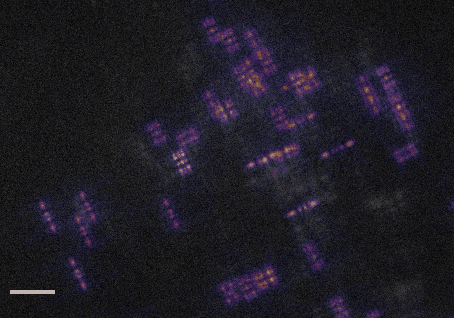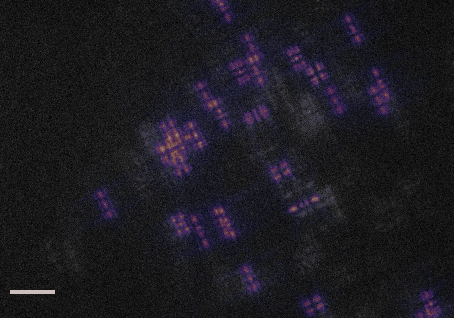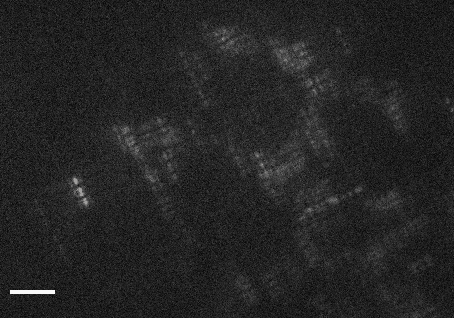
This code accompanies the paper: "DeepSTORM3D: dense 3D localization microcopy and PSF design by deep learning"
- Overview
- System requirements
- Installation instructions
- Code structure
- Demo examples
- Learning a localization model
- Citation
- License
- Contact
This code implements two different applications of CNNs in dense 3D localization microscopy:
- Learning a 3D localization CNN for a given fixed PSF (Tetrapod in this repository).
- Learning an optimized PSF for high density localization via end-to-end optimization.
There's no need to download any dataset as the code itself generates the training and the test sets. Demo 1 illustrates how to train a localization model based on a retreived phase mask, and demo 4 illustrates how the method can be sued to learn an optimzied phase mask. The remaining demos evaluates pre-trained models on both simulated and experimental data.
- The software was tested on a Linux system with Ubuntu version 18.0, and a Windows system with Windows 10 Home.
- Training and evaluation were run on a standard workstation equipped with 32 GB of memory, an Intel(R) Core(TM) i7 − 8700, 3.20 GHz CPU, and a NVidia GeForce Titan Xp GPU with 12 GB of video memory.
- Download this repository as a zip file (or clone it using git).
- Go to the downloaded directory and unzip it.
- The conda environment for this project is given in
environment_<os>.ymlwhere<os>should be substituted with your operating system. For example, to replicate the environment on a linux system use the command:conda env create -f environment_linux.ymlfrom within the downloaded directory. This should take a couple of minutes. - After activation of the environment using:
conda activate deep-storm3d, you're set to go!
- Data generation
DeepSTORM3D/physics_utils.pyimplements the forward physical model relying on Fourier optics.DeepSTORM3D/GeneratingTrainingExamples.pygenerates the training examples (either images + 3D locations as in demo1 or only 3D locations + intensities as in demo4). The assumed physical setup parameters are given in the scriptDemos/parameter_setting_demo1.py. This script should be duplicated and altered according to the experimental setup as detailed inDocs/demo1_documentation.pdf.DeepSTORM3D/data_utils.pyimplements the positions and photons sampling, and defines the dataset classes.- The folder
Mat_Filesincludes phase masks needed to run the demos.
- CNN models and loss function
DeepSTORM3D/cnn_utils.pythis script contains the two CNN models used in this work.DeepSTORM3D/loss_utils.pyimplements the loss function, and an approximation of the Jaccard index.
- Training scripts
DeepSTORM3D/Training_Localization_Model.pythis script trains a localization model based on the pre-calculated training and validation examples inGeneratingTrainingExamples.py. Here, the phase mask is assumed to be fixed (either off-the-shelf or learned), and we're only interested in a dense localization model.DeepSTORM3D/PSF_Learning.pythis script learns an optimized PSF. The training examples in this case are only simulated 3D locations and intensities.
- Post-processing and evaluation
DeepSTORM3D/postprocessing_utils.pyimplements 3D local maxima finding and CoG estimation with a fixed spherical diameter on GPU using max-pooling.DeepSTORM3D/Testing_Localization_Model.pyevaluates a learned localization model on either simulated or experimental images. In demo2/demo5 this module is used with pre-trained weights to localize experimental data. In demo3 it is used to localize simulated data.DeepSTORM3D/assessment_utils.py- this script contains a function that calculates the Jaccard index and the RMSE in both the axial and lateral dimensions given two sets of points in 3D.
- Visualization and saving/loading
DeepSTORM3D/vis_utils.pyincludes plotting functions.DeepSTORM3D/helper_utils.pyincludes saving/loading functions.
-
There are 5 different demo scripts that demonstrate the use of this code:
demo1.py- learns a CNN for localizing high-density Tetrapods under STORM conditions. The script simulates training examples before learning starts. It takes approximately 30 hours to train a model from scratch on a Titan Xp.demo2.py- evaluates a pre-trained CNN for localizing experimental Tetrapods (Fig. 3 main text). The script plots the input images with the localizations voerlaid as red crosses on top. The resulting localizations are saved in a csv file under the folderExperimental_Data/Tetrapod_demo2/. This demo takes about 1 minute on a Titan Xp.demo3.py- evaluates a pre-trained CNN for localizing simulated Tetrapods (Fig. 4 main text). The script plots the simulated input and the regenerated image, and also compares the recovery with the GT positons in 3D. This demo takes about 1 second on a Titan Xp.demo4.py- learns an optimized PSF from scratch. The learned phase mask and its corresponding PSF are plotted each 5 batches in the first 4 epochs, and afterwards only once each 50 batches. Learning takes approximately 30 hours to converge on a Titan Xp.demo5.py- evaluates a pre-trained CNN for localizing an experimental snapshot of a U2OS cell nucleus with the learned PSF. The experimental image can be switched from 'frm1' to 'frm2' inExperimental_Data/. This demo takes about 1 second on a Titan Xp.
-
The
Demosfolder includes the following:Results_Tetrapod_demo2contains pre-trained model weights and training metrics needed to run demo2.Results_Tetrapod_demo3contains pre-trained model weights and training metrics needed to run demo3.Results_Learned_demo5contains pre-trained model weights and training metrics needed to run demo5.parameter_setting_demo*contains the specified setup parameters for each of the demos.
-
The
Experimental_datafolder includes the following:Tetrapod_demo2contains 50 experimental frames from our STORM experiment (Fig. 3 main text).Learned_demo5_frm*contains two snapshots of a U2OS cell nucleus with the learned PSF.
To learn a localization model for your setup, you need to supply a calibrated phase mask (e.g. using beads on the coverslip), and generate a new parameter settings script similar to the ones in the Demos folder. The Docs folder includes the pdf file demo1_documentation.pdf with snapshots detailing the steps in demo1.py to ease the user introduction to DeepSTORM3D. Please go through these while trying demo1.py to get a grasp of how the code works.
If you use this code for your research, please cite our paper:
@article{nehme2020deepstorm3d,
title={DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning},
author={Nehme, Elias and Freedman, Daniel and Gordon, Racheli and Ferdman, Boris and Weiss, Lucien E and Alalouf, Onit and Naor, Tal and Orange, Reut and Michaeli, Tomer and Shechtman, Yoav},
journal={Nature Methods},
volume={17},
number={7},
pages={734--740},
year={2020},
publisher={Nature Publishing Group}
}
This project is covered under the MIT License.
To report any bugs, suggest improvements, or ask questions, please contact me at "seliasne@gmail.com"